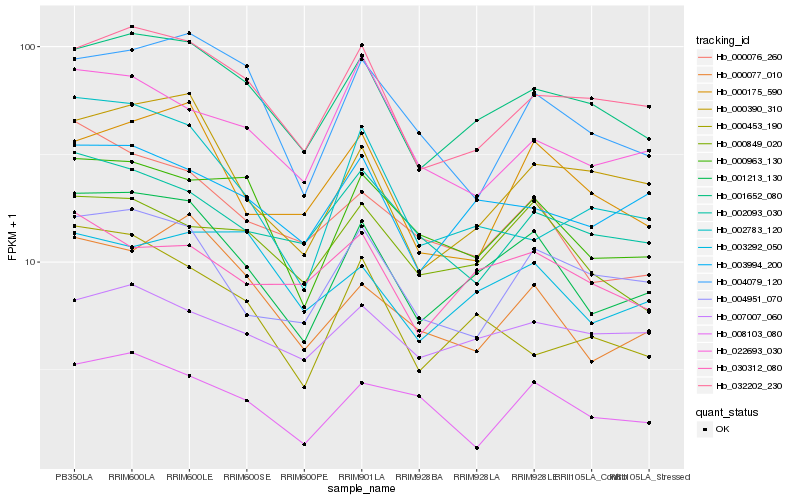
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001213_130 |
0.0 |
- |
- |
PREDICTED: fructokinase-like 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001652_080 |
0.0922382289 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 3 |
Hb_000963_130 |
0.109036114 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000390_310 |
0.1110005304 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000076_260 |
0.1119563704 |
- |
- |
hypothetical protein GUITHDRAFT_149979 [Guillardia theta CCMP2712] |
| 6 |
Hb_032202_230 |
0.11292537 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 7 |
Hb_000077_010 |
0.1133780182 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_003994_200 |
0.1147230509 |
- |
- |
PREDICTED: uncharacterized protein LOC105646142 [Jatropha curcas] |
| 9 |
Hb_004951_070 |
0.114928595 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 10 |
Hb_000849_020 |
0.1222263235 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000175_590 |
0.1260147078 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002783_120 |
0.1275001103 |
- |
- |
PREDICTED: uncharacterized protein LOC105635329 [Jatropha curcas] |
| 13 |
Hb_000453_190 |
0.128779833 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 14 |
Hb_004079_120 |
0.1309408804 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 15 |
Hb_003292_050 |
0.132941504 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 16 |
Hb_030312_080 |
0.1341913755 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 17 |
Hb_002093_030 |
0.1356966105 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 18 |
Hb_007007_060 |
0.1358348581 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_022693_030 |
0.1363876607 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 20 |
Hb_008103_080 |
0.1369087805 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |