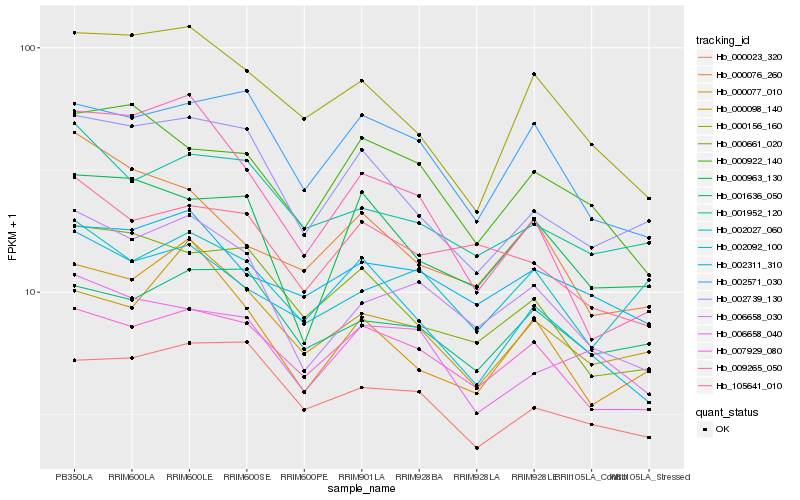
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006658_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_04047 [Jatropha curcas] |
| 2 |
Hb_000077_010 |
0.0991547767 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_001952_120 |
0.1063538803 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_105641_010 |
0.1083929324 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 5 |
Hb_007929_080 |
0.1105641918 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 6 |
Hb_000661_020 |
0.1138831553 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 7 |
Hb_002311_310 |
0.1141733758 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 8 |
Hb_009265_050 |
0.1159871054 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 9 |
Hb_002739_130 |
0.1163993024 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 10 |
Hb_000156_160 |
0.116588799 |
- |
- |
PREDICTED: urease accessory protein G [Jatropha curcas] |
| 11 |
Hb_006658_040 |
0.1174555063 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |
| 12 |
Hb_000023_320 |
0.1175613435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000963_130 |
0.1183088532 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_002027_060 |
0.1216756441 |
- |
- |
PREDICTED: exosome complex component RRP4 [Jatropha curcas] |
| 15 |
Hb_002571_030 |
0.1217701522 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 16 |
Hb_000076_260 |
0.1229304769 |
- |
- |
hypothetical protein GUITHDRAFT_149979 [Guillardia theta CCMP2712] |
| 17 |
Hb_000922_140 |
0.1239078993 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 18 |
Hb_000098_140 |
0.124818667 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 19 |
Hb_002092_100 |
0.1263942874 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 20 |
Hb_001636_050 |
0.1272076986 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |