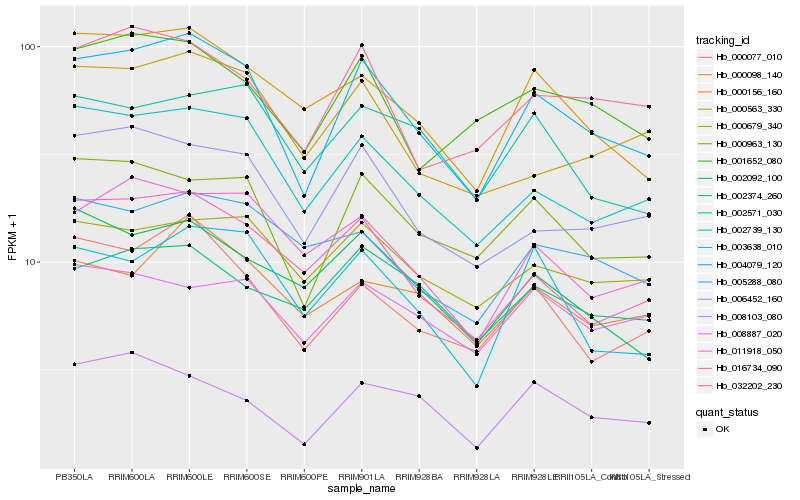
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004079_120 |
0.0 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 2 |
Hb_000963_130 |
0.082555524 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_002739_130 |
0.0851359618 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 4 |
Hb_006452_160 |
0.0951637041 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 5 |
Hb_000077_010 |
0.0966205395 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000156_160 |
0.0988408908 |
- |
- |
PREDICTED: urease accessory protein G [Jatropha curcas] |
| 7 |
Hb_016734_090 |
0.1011559349 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 8 |
Hb_032202_230 |
0.1013222217 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 9 |
Hb_001652_080 |
0.1050145856 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 10 |
Hb_008103_080 |
0.1056123366 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_011918_050 |
0.106380419 |
- |
- |
- |
| 12 |
Hb_000679_340 |
0.1075448543 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003638_010 |
0.1099056268 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |
| 14 |
Hb_000563_330 |
0.110447946 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_002092_100 |
0.1129708367 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 16 |
Hb_000098_140 |
0.1134414991 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 17 |
Hb_002374_260 |
0.1141023789 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 18 |
Hb_002571_030 |
0.1142733057 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 19 |
Hb_005288_080 |
0.115904398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008887_020 |
0.117623269 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |