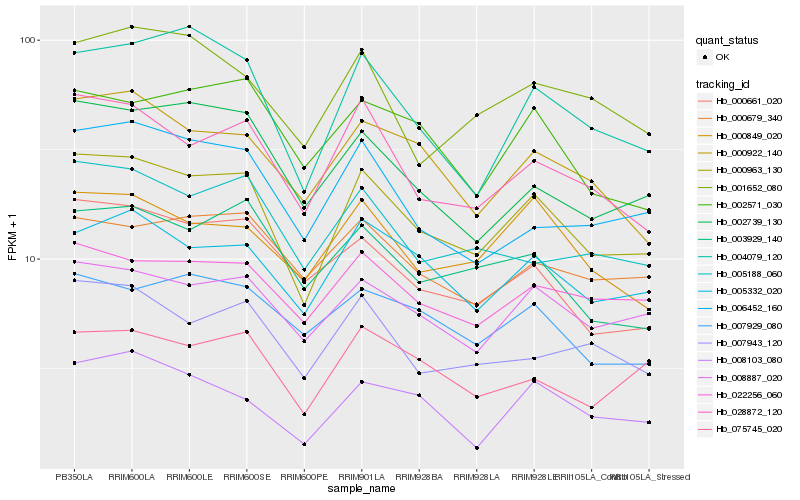
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000963_130 |
0.0 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_028872_120 |
0.0783508724 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 3 |
Hb_005332_020 |
0.0813708788 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_008887_020 |
0.0825293162 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 5 |
Hb_004079_120 |
0.082555524 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 6 |
Hb_000922_140 |
0.0908966999 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 7 |
Hb_022256_060 |
0.0915389569 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 8 |
Hb_000849_020 |
0.0945119698 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002739_130 |
0.0946913552 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 10 |
Hb_007943_120 |
0.0953202921 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 11 |
Hb_075745_020 |
0.0961133242 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003929_140 |
0.0962054905 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_005188_060 |
0.0965491086 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 14 |
Hb_000661_020 |
0.0973304057 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 15 |
Hb_006452_160 |
0.0988786284 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 16 |
Hb_008103_080 |
0.0996025856 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000679_340 |
0.0996431188 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007929_080 |
0.0996786317 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 19 |
Hb_001652_080 |
0.1000419326 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 20 |
Hb_002571_030 |
0.1005180943 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |