| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
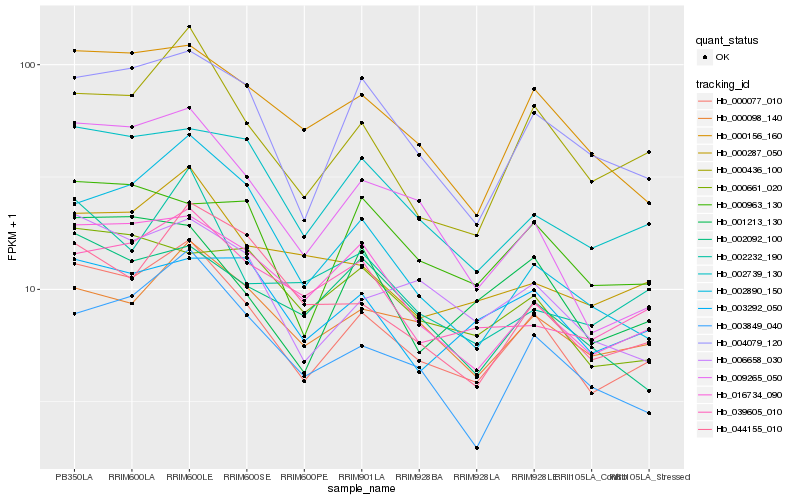
Hb_000077_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000436_100 |
0.0891932333 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 3 |
Hb_016734_090 |
0.0953454151 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 4 |
Hb_002739_130 |
0.0958999482 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 5 |
Hb_004079_120 |
0.0966205395 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 6 |
Hb_000156_160 |
0.0982850362 |
- |
- |
PREDICTED: urease accessory protein G [Jatropha curcas] |
| 7 |
Hb_006658_030 |
0.0991547767 |
- |
- |
hypothetical protein JCGZ_04047 [Jatropha curcas] |
| 8 |
Hb_003849_040 |
0.1039733624 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 9 |
Hb_000098_140 |
0.1049595007 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 10 |
Hb_009265_050 |
0.1057303869 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 11 |
Hb_000287_050 |
0.1113295121 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 12 |
Hb_001213_130 |
0.1133780182 |
- |
- |
PREDICTED: fructokinase-like 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_039605_010 |
0.1143437183 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002890_150 |
0.1167535592 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 15 |
Hb_044155_010 |
0.1168797142 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000661_020 |
0.1184805123 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 17 |
Hb_002232_190 |
0.1192359752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002092_100 |
0.119538471 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 19 |
Hb_003292_050 |
0.1210271863 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 20 |
Hb_000963_130 |
0.1228506414 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |