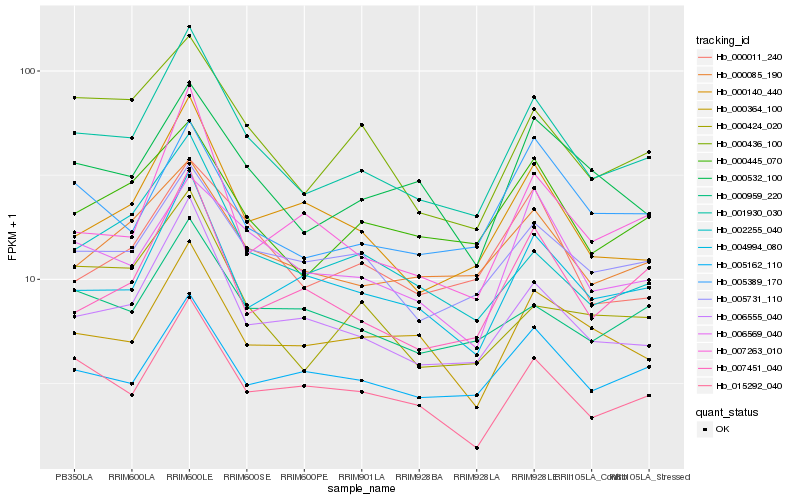
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001930_030 |
0.0 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 2 |
Hb_006555_040 |
0.0772657968 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 3 |
Hb_005731_110 |
0.0950754571 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 4 |
Hb_000445_070 |
0.0964889979 |
- |
- |
PREDICTED: uncharacterized protein LOC105638998 [Jatropha curcas] |
| 5 |
Hb_015292_040 |
0.0984807126 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 6 |
Hb_004994_080 |
0.0989828675 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000436_100 |
0.1006327587 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 8 |
Hb_000085_190 |
0.1024164651 |
- |
- |
FtsZ1 protein [Manihot esculenta] |
| 9 |
Hb_000532_100 |
0.1052031015 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 10 |
Hb_000140_440 |
0.1059110898 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 11 |
Hb_002255_040 |
0.1072393175 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 12 |
Hb_007451_040 |
0.1090101341 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 13 |
Hb_006569_040 |
0.1095164241 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 14 |
Hb_005162_110 |
0.1105766705 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 15 |
Hb_000959_220 |
0.110846221 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_005389_170 |
0.1131999258 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 17 |
Hb_000364_100 |
0.1134494019 |
- |
- |
PREDICTED: uncharacterized protein LOC105639739 [Jatropha curcas] |
| 18 |
Hb_007263_010 |
0.1138667506 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 19 |
Hb_000011_240 |
0.1149926214 |
- |
- |
PREDICTED: uncharacterized protein LOC105631316 [Jatropha curcas] |
| 20 |
Hb_000424_020 |
0.1158985555 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |