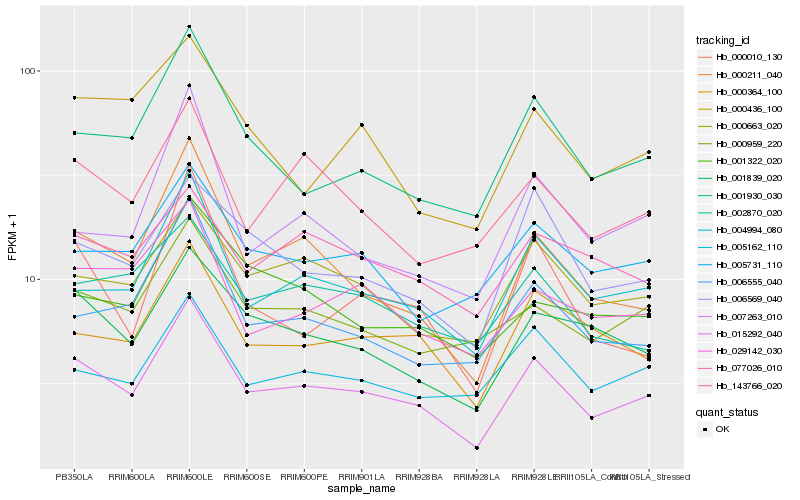
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015292_040 |
0.0 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 2 |
Hb_004994_080 |
0.0897404028 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000211_040 |
0.0899404046 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 4 |
Hb_001930_030 |
0.0984807126 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000663_020 |
0.1026968838 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_001839_020 |
0.1034893447 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 7 |
Hb_000364_100 |
0.1043514546 |
- |
- |
PREDICTED: uncharacterized protein LOC105639739 [Jatropha curcas] |
| 8 |
Hb_000959_220 |
0.1044741202 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_006555_040 |
0.1073563549 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 10 |
Hb_143766_020 |
0.1093190035 |
- |
- |
putative phospholipase A2 [Gossypium arboreum] |
| 11 |
Hb_029142_030 |
0.1101164502 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 12 |
Hb_005162_110 |
0.1114746906 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 13 |
Hb_005731_110 |
0.113124078 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 14 |
Hb_006569_040 |
0.1145354318 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 15 |
Hb_002870_020 |
0.1159230852 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000436_100 |
0.1211251739 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 17 |
Hb_001322_020 |
0.1214802559 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 18 |
Hb_007263_010 |
0.1237143775 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 19 |
Hb_000010_130 |
0.1237193578 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 11, chloroplastic [Pyrus x bretschneideri] |
| 20 |
Hb_077026_010 |
0.1244966108 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |