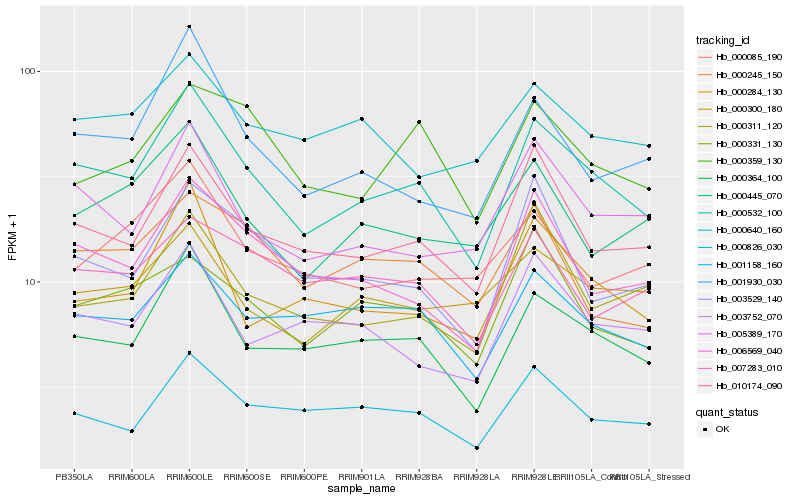
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000532_100 |
0.0 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 2 |
Hb_000364_100 |
0.0719236388 |
- |
- |
PREDICTED: uncharacterized protein LOC105639739 [Jatropha curcas] |
| 3 |
Hb_010174_090 |
0.0894711681 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_006569_040 |
0.1003669591 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 5 |
Hb_005389_170 |
0.1046041111 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 6 |
Hb_001930_030 |
0.1052031015 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000826_030 |
0.1052537924 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 8 |
Hb_001158_160 |
0.1066965174 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000445_070 |
0.1133172993 |
- |
- |
PREDICTED: uncharacterized protein LOC105638998 [Jatropha curcas] |
| 10 |
Hb_000284_130 |
0.1136180504 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 11 |
Hb_003529_140 |
0.1163001344 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003752_070 |
0.1174727445 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000311_120 |
0.1201864733 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 14 |
Hb_000331_130 |
0.120392139 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000300_180 |
0.1211554992 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 16 |
Hb_000640_160 |
0.1225635015 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 17 |
Hb_000359_130 |
0.1227589876 |
- |
- |
4-hydroxyphenylpyruvate dioxygenase [Hevea brasiliensis] |
| 18 |
Hb_000245_150 |
0.1240495924 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 19 |
Hb_007283_010 |
0.1244558894 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 20 |
Hb_000085_190 |
0.1251534091 |
- |
- |
FtsZ1 protein [Manihot esculenta] |