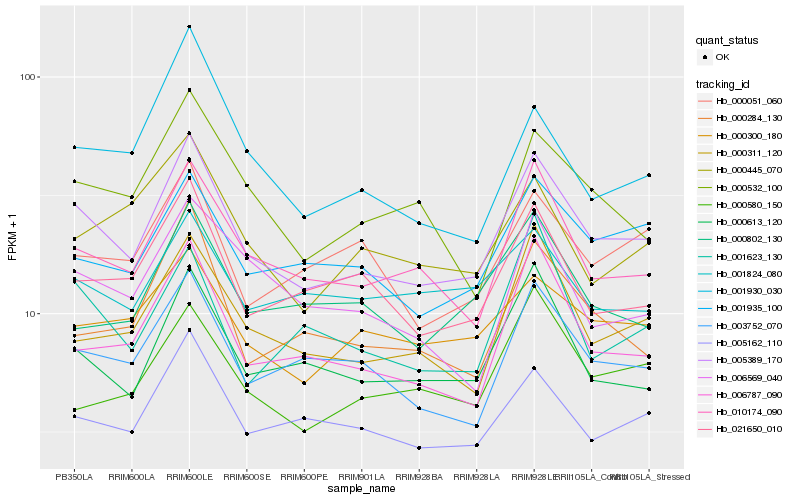
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005389_170 |
0.0 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 2 |
Hb_010174_090 |
0.0818777906 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_003752_070 |
0.0892834361 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000613_120 |
0.0902846408 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005162_110 |
0.0907713025 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 6 |
Hb_001935_100 |
0.0954576958 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 7 |
Hb_006787_090 |
0.0958000682 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 8 |
Hb_000311_120 |
0.0959894233 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 9 |
Hb_021650_010 |
0.0995421738 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 10 |
Hb_000300_180 |
0.1034551502 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 11 |
Hb_000532_100 |
0.1046041111 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 12 |
Hb_000284_130 |
0.1082251708 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 13 |
Hb_000051_060 |
0.1104851772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006569_040 |
0.1117811659 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 15 |
Hb_000580_150 |
0.111919668 |
- |
- |
PREDICTED: probable tRNA N6-adenosine threonylcarbamoyltransferase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001623_130 |
0.1131742327 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_001930_030 |
0.1131999258 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000445_070 |
0.1144106034 |
- |
- |
PREDICTED: uncharacterized protein LOC105638998 [Jatropha curcas] |
| 19 |
Hb_001824_080 |
0.1153393033 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 20 |
Hb_000802_130 |
0.1161317572 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |