| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
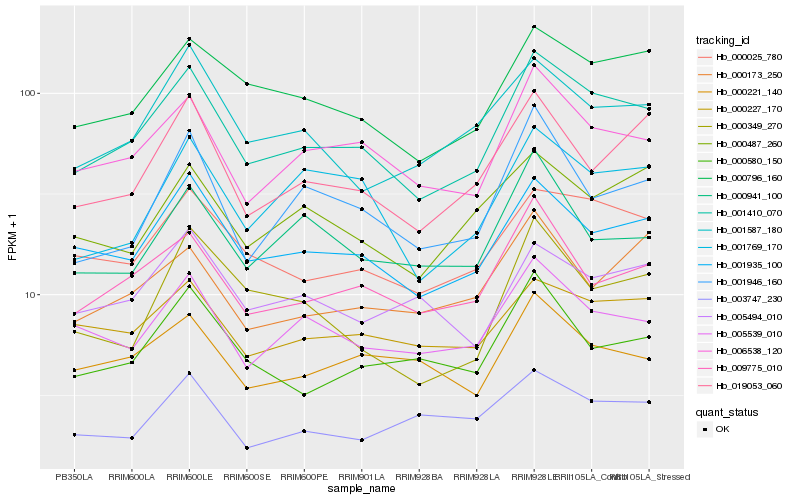
Hb_001410_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 2 |
Hb_006538_120 |
0.0733052742 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001935_100 |
0.0767013137 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 4 |
Hb_000796_160 |
0.0793113958 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000025_780 |
0.0803902051 |
- |
- |
PREDICTED: sanguinarine reductase [Jatropha curcas] |
| 6 |
Hb_009775_010 |
0.0890844648 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 7 |
Hb_005539_010 |
0.0961466657 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 8 |
Hb_019053_060 |
0.0965331388 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 9 |
Hb_000227_170 |
0.1003875202 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 10 |
Hb_001587_180 |
0.1004413681 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 11 |
Hb_001769_170 |
0.1018435878 |
- |
- |
50S ribosomal protein L31, putative [Ricinus communis] |
| 12 |
Hb_001946_160 |
0.1033441401 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 13 |
Hb_000221_140 |
0.1060363577 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000580_150 |
0.1068479136 |
- |
- |
PREDICTED: probable tRNA N6-adenosine threonylcarbamoyltransferase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000487_260 |
0.1073233401 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005494_010 |
0.1107055197 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_000941_100 |
0.1115813505 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 18 |
Hb_000173_250 |
0.114689777 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 19 |
Hb_000349_270 |
0.1161665581 |
- |
- |
PREDICTED: uncharacterized protein ycf49 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003747_230 |
0.116482124 |
- |
- |
PREDICTED: uncharacterized protein LOC105630239 isoform X1 [Jatropha curcas] |