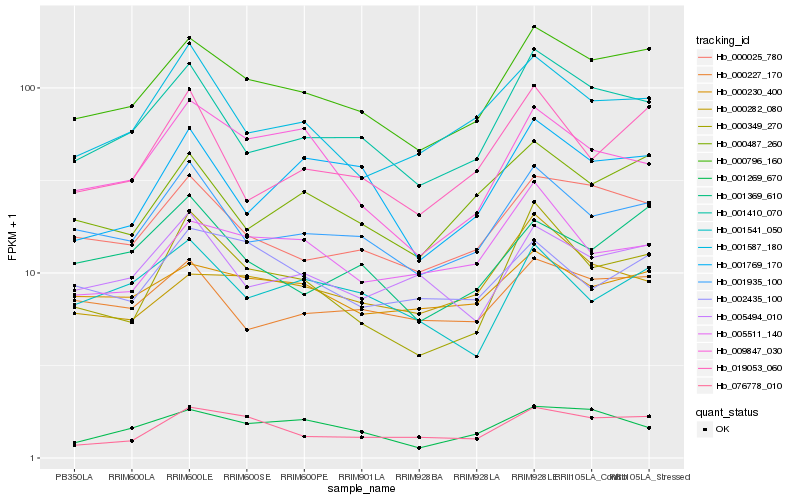
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000796_160 |
0.0 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000025_780 |
0.0741040015 |
- |
- |
PREDICTED: sanguinarine reductase [Jatropha curcas] |
| 3 |
Hb_001935_100 |
0.0789987576 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 4 |
Hb_001410_070 |
0.0793113958 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 5 |
Hb_000487_260 |
0.0876368884 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 6 |
Hb_076778_010 |
0.0916738896 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 7 |
Hb_000230_400 |
0.0998026463 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000227_170 |
0.0998999435 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_005494_010 |
0.1026856333 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_001369_610 |
0.1035746429 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000349_270 |
0.1035921132 |
- |
- |
PREDICTED: uncharacterized protein ycf49 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001587_180 |
0.1039499875 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 13 |
Hb_019053_060 |
0.1062287945 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 14 |
Hb_001269_670 |
0.1072681038 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 15 |
Hb_005511_140 |
0.1110697104 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 16 |
Hb_001769_170 |
0.1112387142 |
- |
- |
50S ribosomal protein L31, putative [Ricinus communis] |
| 17 |
Hb_000282_080 |
0.112204069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002435_100 |
0.1139044704 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 19 |
Hb_001541_050 |
0.1142720253 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 20 |
Hb_009847_030 |
0.1152191776 |
- |
- |
zinc finger family protein [Populus trichocarpa] |