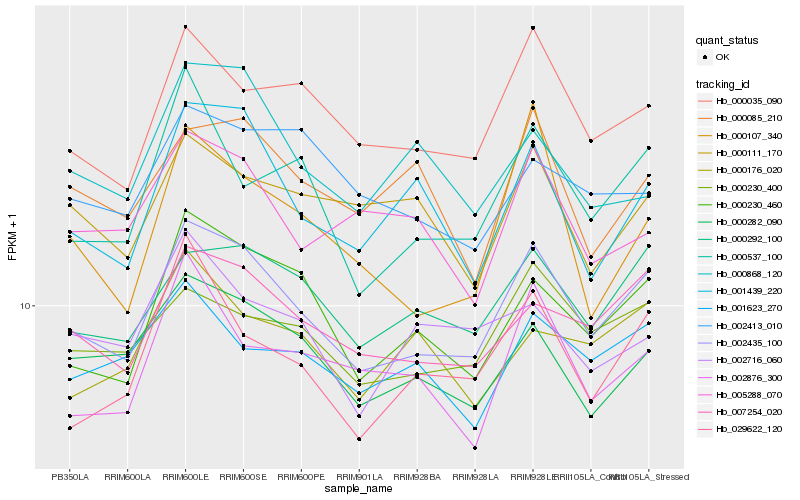
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002435_100 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 2 |
Hb_007254_020 |
0.0627133708 |
- |
- |
PREDICTED: uncharacterized protein LOC105629137 [Jatropha curcas] |
| 3 |
Hb_000292_100 |
0.067638506 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 4 |
Hb_000230_460 |
0.0690887943 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 5 |
Hb_000035_090 |
0.0707731548 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 6 |
Hb_000230_400 |
0.0773835667 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000282_090 |
0.0785523621 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 8 |
Hb_000537_100 |
0.0833234636 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001439_220 |
0.0833681302 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 10 |
Hb_029622_120 |
0.0855202864 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 11 |
Hb_001623_270 |
0.0886924602 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 12 |
Hb_000176_020 |
0.0936227982 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 13 |
Hb_002876_300 |
0.0943012251 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000111_170 |
0.0955095252 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000085_210 |
0.0960585613 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 16 |
Hb_000107_340 |
0.0980550201 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 17 |
Hb_005288_070 |
0.0984117914 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_002413_010 |
0.0984661799 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_002716_060 |
0.0997155322 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 20 |
Hb_000868_120 |
0.1001909652 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |