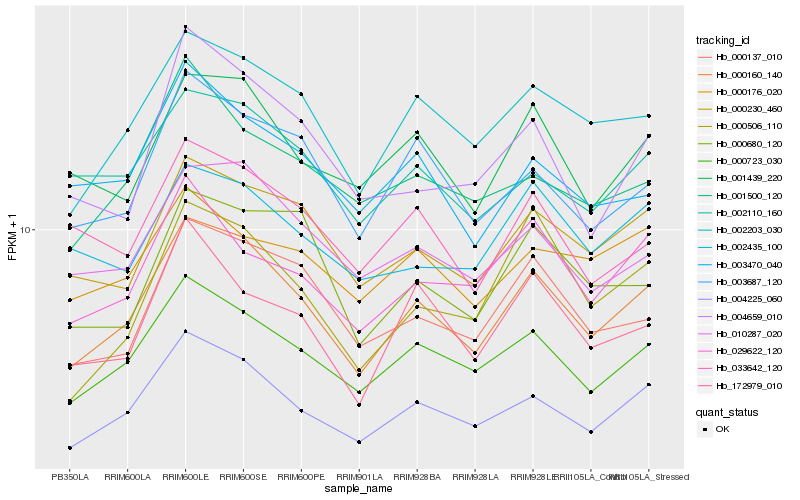
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000160_140 |
0.0 |
- |
- |
PREDICTED: ELL-associated factor 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_004225_060 |
0.0499234432 |
- |
- |
hypothetical protein CISIN_1g027511mg [Citrus sinensis] |
| 3 |
Hb_000723_030 |
0.0601546178 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X1 [Jatropha curcas] |
| 4 |
Hb_002203_030 |
0.0677765085 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 5 |
Hb_000230_460 |
0.0795328723 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 6 |
Hb_010287_020 |
0.085134997 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 7 |
Hb_000137_010 |
0.0852651414 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 8 |
Hb_001439_220 |
0.0863959216 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 9 |
Hb_003687_120 |
0.0878505036 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 10 |
Hb_029622_120 |
0.0958738168 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 11 |
Hb_002110_160 |
0.0959879243 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_172979_010 |
0.0969209869 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 13 |
Hb_000506_110 |
0.0998131181 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 14 |
Hb_000680_120 |
0.1004597423 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 15 |
Hb_033642_120 |
0.1011240482 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 16 |
Hb_003470_040 |
0.1018659658 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_002435_100 |
0.1021891888 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 18 |
Hb_001500_120 |
0.1025252431 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000176_020 |
0.1033197991 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 20 |
Hb_004659_010 |
0.1034249238 |
- |
- |
Guanine nucleotide-binding subunit beta-2 [Gossypium arboreum] |