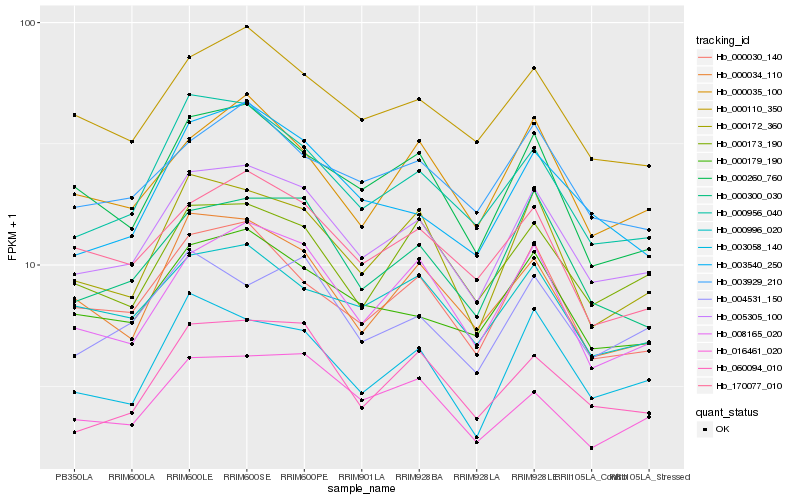
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005305_100 |
0.0 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000956_040 |
0.0615437489 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 3 |
Hb_000300_030 |
0.0624005888 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 4 |
Hb_000172_360 |
0.0643167233 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 5 |
Hb_008165_020 |
0.0660470955 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000179_190 |
0.0667052629 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 7 |
Hb_000110_350 |
0.0686575027 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_000030_140 |
0.0688067133 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003058_140 |
0.0710351088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_060094_010 |
0.0713765784 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000260_760 |
0.0714276027 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 12 |
Hb_000173_190 |
0.0722976688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003929_210 |
0.0755101516 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000034_110 |
0.0755649669 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 15 |
Hb_000035_100 |
0.0763588397 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_003540_250 |
0.0769064707 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 17 |
Hb_016461_020 |
0.0771691863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000996_020 |
0.0777231252 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 19 |
Hb_170077_010 |
0.0783225018 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004531_150 |
0.0784306557 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |