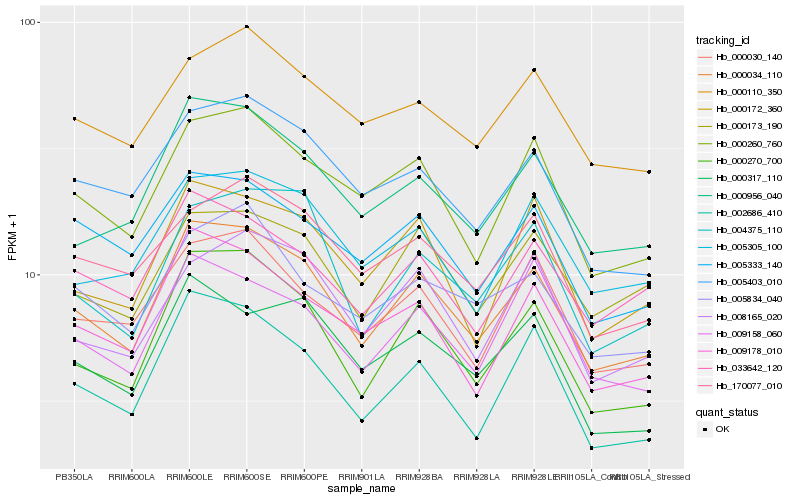
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000034_110 |
0.0 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 2 |
Hb_000270_700 |
0.0539312274 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 3 |
Hb_000956_040 |
0.0651490856 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 4 |
Hb_000260_760 |
0.065562154 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 5 |
Hb_009178_010 |
0.0678622123 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 6 |
Hb_002686_410 |
0.0704643339 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_005403_010 |
0.072868007 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 8 |
Hb_000030_140 |
0.0749090587 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000172_360 |
0.0751454005 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 10 |
Hb_005305_100 |
0.0755649669 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000173_190 |
0.0765436148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_033642_120 |
0.0772207788 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 13 |
Hb_004375_110 |
0.0778515125 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 14 |
Hb_005333_140 |
0.0788784092 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009158_060 |
0.0794408475 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 16 |
Hb_000110_350 |
0.0801605895 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 17 |
Hb_170077_010 |
0.0802480986 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008165_020 |
0.0804460463 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000317_110 |
0.0806837361 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 20 |
Hb_005834_040 |
0.0817409636 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |