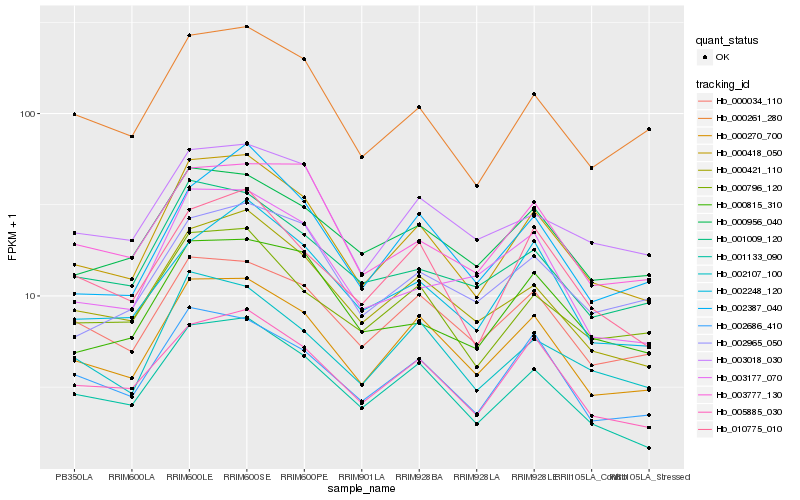
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000418_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 2 |
Hb_000270_700 |
0.0586935068 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 3 |
Hb_001133_090 |
0.064890977 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 4 |
Hb_000421_110 |
0.0678585073 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005885_030 |
0.074453551 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 6 |
Hb_000261_280 |
0.0770274344 |
- |
- |
BnaA10g23810D [Brassica napus] |
| 7 |
Hb_002107_100 |
0.0805029809 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 8 |
Hb_000796_120 |
0.0814759032 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_002686_410 |
0.0822010767 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_010775_010 |
0.0832252023 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000815_310 |
0.0860045092 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001009_120 |
0.0890785396 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002387_040 |
0.0893150271 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 14 |
Hb_000956_040 |
0.090033908 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 15 |
Hb_000034_110 |
0.0903131793 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 16 |
Hb_003018_030 |
0.0903597707 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 17 |
Hb_002248_120 |
0.0929849299 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 18 |
Hb_003177_070 |
0.0935367968 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 19 |
Hb_003777_130 |
0.0943819351 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 20 |
Hb_002965_050 |
0.0966110322 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |