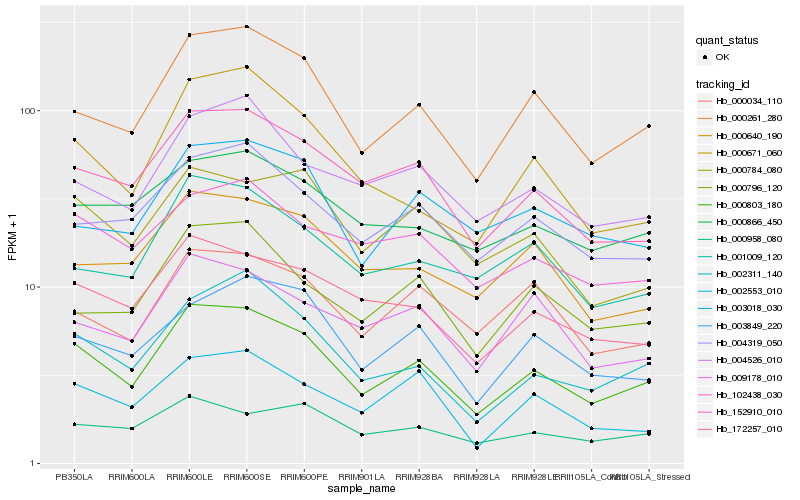
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_180 |
0.0 |
- |
- |
hexokinase [Manihot esculenta] |
| 2 |
Hb_000261_280 |
0.0717180965 |
- |
- |
BnaA10g23810D [Brassica napus] |
| 3 |
Hb_152910_010 |
0.087413412 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 4 |
Hb_002311_140 |
0.0936222438 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 5 |
Hb_172257_010 |
0.0958082695 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 6 |
Hb_004319_050 |
0.1012515294 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 7 |
Hb_003849_220 |
0.1063589008 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 8 |
Hb_000796_120 |
0.1068069579 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_004526_010 |
0.1075769056 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |
| 10 |
Hb_102438_030 |
0.1086638082 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000784_080 |
0.1099133922 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 12 |
Hb_000640_190 |
0.110340903 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001009_120 |
0.1107739602 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002553_010 |
0.1124426658 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 2-like [Populus euphratica] |
| 15 |
Hb_000866_450 |
0.1128425475 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_000034_110 |
0.1131437189 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 17 |
Hb_003018_030 |
0.1133654204 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 18 |
Hb_009178_010 |
0.113583965 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 19 |
Hb_000958_080 |
0.1144167834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000671_060 |
0.1148849863 |
- |
- |
protein binding protein, putative [Ricinus communis] |