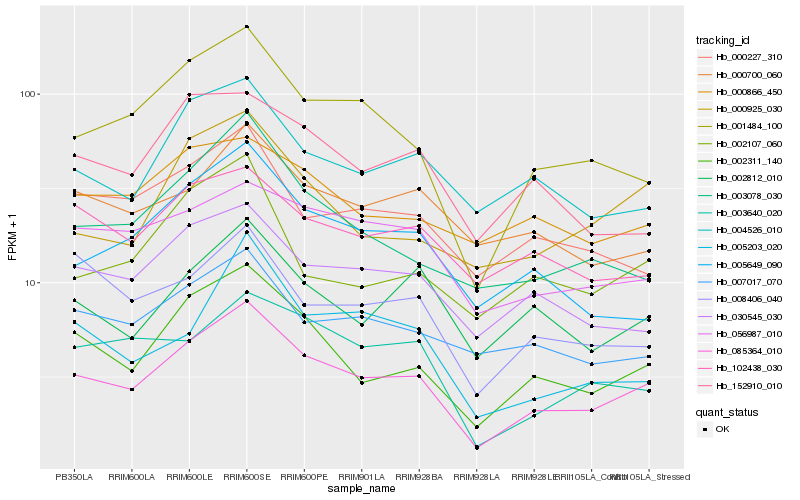
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_085364_010 |
0.0 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 2 |
Hb_002311_140 |
0.0886128737 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 3 |
Hb_001484_100 |
0.102363742 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 4 |
Hb_030545_030 |
0.1181447399 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 5 |
Hb_003078_030 |
0.1198462701 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 6 |
Hb_004526_010 |
0.122312846 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |
| 7 |
Hb_002812_010 |
0.1238556801 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_056987_010 |
0.1253381957 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 9 |
Hb_005649_090 |
0.1257499997 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 10 |
Hb_003640_020 |
0.1266768273 |
- |
- |
pelota, putative [Ricinus communis] |
| 11 |
Hb_000227_310 |
0.1279163349 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_007017_070 |
0.1288603199 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000925_030 |
0.1302366862 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 14 |
Hb_102438_030 |
0.130715644 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_152910_010 |
0.1309332216 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 16 |
Hb_000700_060 |
0.1316477149 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 17 |
Hb_008406_040 |
0.1331840045 |
- |
- |
- |
| 18 |
Hb_005203_020 |
0.1344937682 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 19 |
Hb_000866_450 |
0.1362385191 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_002107_060 |
0.1363781139 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |