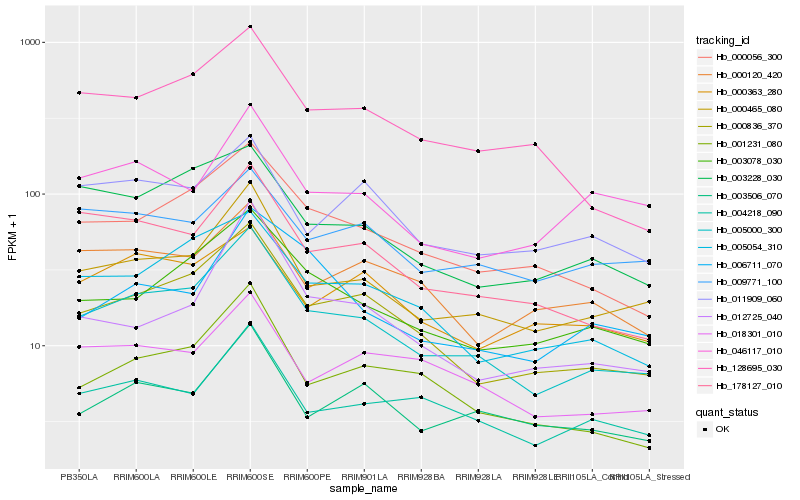
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005000_300 |
0.0 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 2 |
Hb_000836_370 |
0.072644327 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000465_080 |
0.0742970725 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 4 |
Hb_000056_300 |
0.0893846128 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003078_030 |
0.0953181873 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 6 |
Hb_001231_080 |
0.1008267597 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 7 |
Hb_005054_310 |
0.1038723003 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 8 |
Hb_178127_010 |
0.1068559527 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 9 |
Hb_128695_030 |
0.1101089638 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 10 |
Hb_004218_090 |
0.1131361468 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_003506_070 |
0.1199544108 |
- |
- |
PREDICTED: calcineurin B-like protein 7 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003228_030 |
0.1216849499 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_018301_010 |
0.1277331067 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 14 |
Hb_012725_040 |
0.1340838046 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_006711_070 |
0.1355885998 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 16 |
Hb_011909_060 |
0.137101725 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 17 |
Hb_000120_420 |
0.1388319306 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 18 |
Hb_000363_280 |
0.139392812 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 19 |
Hb_009771_100 |
0.1396334842 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 20 |
Hb_046117_010 |
0.1397758952 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |