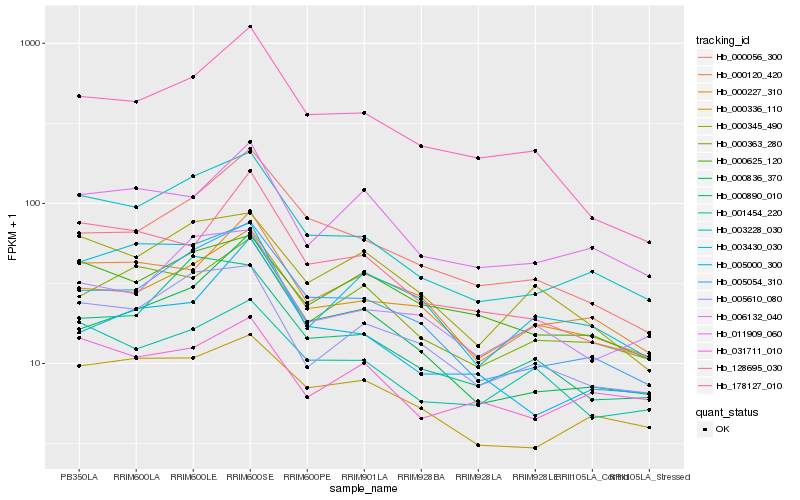
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003228_030 |
0.0 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_005054_310 |
0.0743185631 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 3 |
Hb_000336_110 |
0.095246658 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_005610_080 |
0.0953683384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000890_010 |
0.1060842849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006132_040 |
0.1071269226 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 7 |
Hb_000227_310 |
0.1073834643 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_001454_220 |
0.1114110445 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 9 |
Hb_000345_490 |
0.1135935914 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 10 |
Hb_000056_300 |
0.114435181 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000120_420 |
0.1153613709 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 12 |
Hb_000363_280 |
0.1165174757 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 13 |
Hb_128695_030 |
0.1170443766 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 14 |
Hb_178127_010 |
0.1172551706 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 15 |
Hb_011909_060 |
0.1188392214 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 16 |
Hb_003430_030 |
0.1190539951 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 17 |
Hb_000836_370 |
0.1192151831 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_031711_010 |
0.1202615921 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000625_120 |
0.1210210563 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 20 |
Hb_005000_300 |
0.1216849499 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |