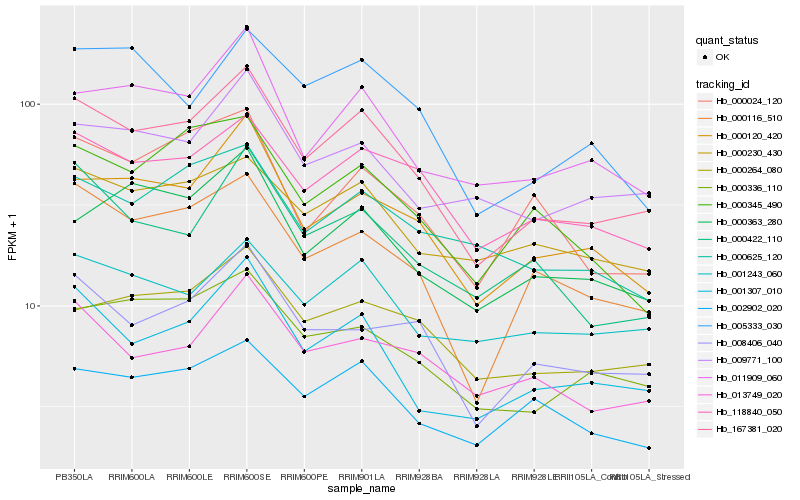
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_167381_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 2 |
Hb_001307_010 |
0.0819395866 |
- |
- |
PREDICTED: uncharacterized protein LOC103939917 [Pyrus x bretschneideri] |
| 3 |
Hb_000116_510 |
0.0862163144 |
- |
- |
PREDICTED: RNA-binding protein 24-A-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000120_420 |
0.0864704275 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 5 |
Hb_118840_050 |
0.089620583 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 6 |
Hb_000336_110 |
0.0922751124 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_011909_060 |
0.0940038028 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 8 |
Hb_008406_040 |
0.0949377713 |
- |
- |
- |
| 9 |
Hb_000264_080 |
0.0979768383 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 10 |
Hb_000422_110 |
0.0991234111 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 11 |
Hb_002902_020 |
0.0994487558 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 12 |
Hb_000230_430 |
0.1006890076 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_013749_020 |
0.1029001397 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_001243_060 |
0.1059586989 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 15 |
Hb_009771_100 |
0.1059595872 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 16 |
Hb_000363_280 |
0.1061126742 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 17 |
Hb_000024_120 |
0.1077395174 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 18 |
Hb_000625_120 |
0.1082760762 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 19 |
Hb_005333_030 |
0.1086015902 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 20 |
Hb_000345_490 |
0.1087618806 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |