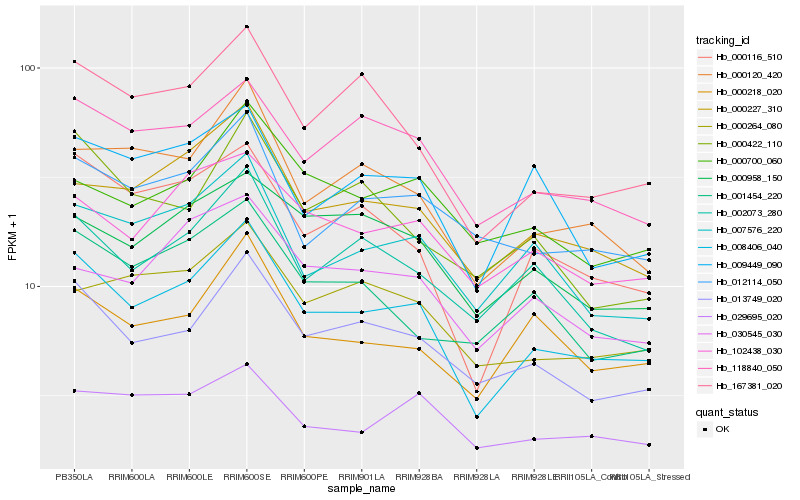
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008406_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_013749_020 |
0.0831962722 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_000116_510 |
0.0880252926 |
- |
- |
PREDICTED: RNA-binding protein 24-A-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000120_420 |
0.0928883954 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 5 |
Hb_167381_020 |
0.0949377713 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 6 |
Hb_000227_310 |
0.0950418357 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_118840_050 |
0.0957536853 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 8 |
Hb_007576_220 |
0.099771702 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 9 |
Hb_002073_280 |
0.1029545157 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000218_020 |
0.1040600451 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 11 |
Hb_102438_030 |
0.1042851413 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_012114_050 |
0.1059673669 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009449_090 |
0.1094797546 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000264_080 |
0.1115927207 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 15 |
Hb_000958_150 |
0.1122572573 |
- |
- |
Actin, putative [Ricinus communis] |
| 16 |
Hb_030545_030 |
0.1143425421 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 17 |
Hb_029695_020 |
0.1146391752 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001454_220 |
0.1147731243 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 19 |
Hb_000422_110 |
0.1148448944 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 20 |
Hb_000700_060 |
0.1157855025 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |