| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
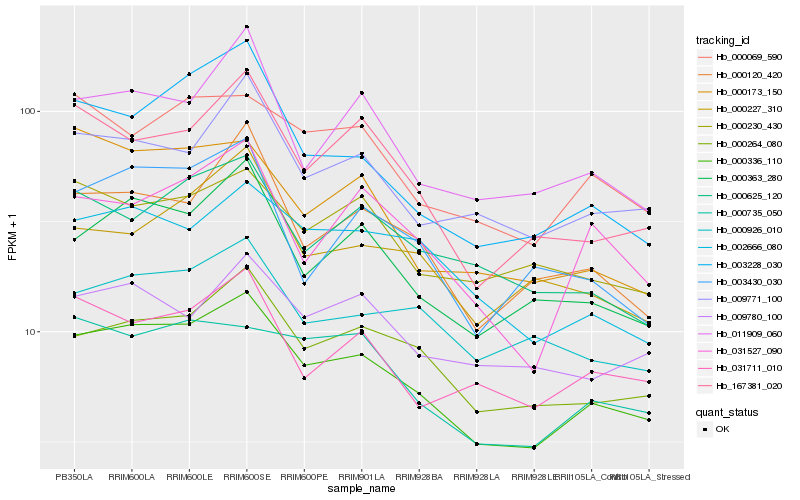
Hb_000336_110 |
0.0 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000264_080 |
0.0762351467 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 3 |
Hb_000363_280 |
0.0805065387 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 4 |
Hb_000120_420 |
0.089950867 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 5 |
Hb_009771_100 |
0.0920903451 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 6 |
Hb_167381_020 |
0.0922751124 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 7 |
Hb_000069_590 |
0.0933924664 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 8 |
Hb_003228_030 |
0.095246658 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_002666_080 |
0.0963246222 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_011909_060 |
0.0964538002 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 11 |
Hb_000735_050 |
0.0979791047 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 12 |
Hb_031711_010 |
0.0997548231 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_031527_090 |
0.1001476488 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003430_030 |
0.1009806797 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 15 |
Hb_000926_010 |
0.1019703422 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 16 |
Hb_000230_430 |
0.102035978 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_000625_120 |
0.1021536936 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 18 |
Hb_000173_150 |
0.1026448382 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 19 |
Hb_000227_310 |
0.1045913425 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_009780_100 |
0.1051685478 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |