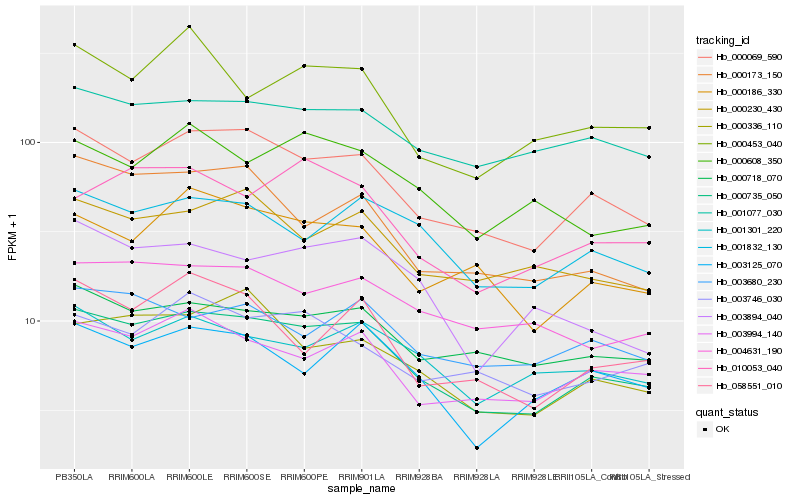
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000735_050 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 2 |
Hb_000069_590 |
0.0542276374 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 3 |
Hb_003994_140 |
0.0893502723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000718_070 |
0.0910696287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000186_330 |
0.0972972928 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 6 |
Hb_000336_110 |
0.0979791047 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_001832_130 |
0.0985948744 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 8 |
Hb_000230_430 |
0.0986046409 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_001301_220 |
0.1003000085 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 10 |
Hb_003125_070 |
0.1015923693 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio1-like [Jatropha curcas] |
| 11 |
Hb_000173_150 |
0.1020770673 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 12 |
Hb_001077_030 |
0.1022143268 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003746_030 |
0.1039192018 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 14 |
Hb_004631_190 |
0.1041168819 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 15 |
Hb_010053_040 |
0.1047466889 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 16 |
Hb_003894_040 |
0.1050670625 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 17 |
Hb_000453_040 |
0.1090077271 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 18 |
Hb_000608_350 |
0.1090786913 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 19 |
Hb_058551_010 |
0.1097272227 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_003680_230 |
0.1097372109 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |