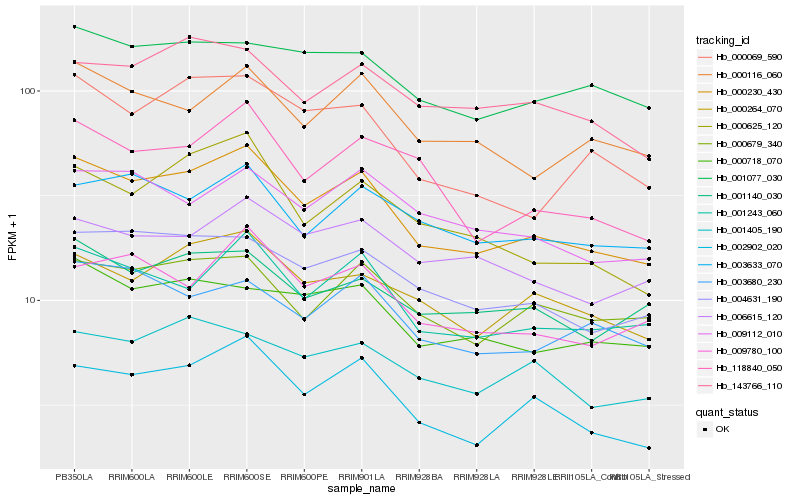
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_430 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_001243_060 |
0.0587289964 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 3 |
Hb_002902_020 |
0.0639657793 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 4 |
Hb_004631_190 |
0.0689490749 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 5 |
Hb_000264_070 |
0.071587568 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 6 |
Hb_009780_100 |
0.0776768133 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 7 |
Hb_118840_050 |
0.0779573937 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 8 |
Hb_000625_120 |
0.0781753997 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 9 |
Hb_000679_340 |
0.0788487899 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000116_060 |
0.0793097395 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 11 |
Hb_000718_070 |
0.0798121289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001140_030 |
0.0822015023 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 13 |
Hb_000069_590 |
0.083079635 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 14 |
Hb_001405_190 |
0.0844554776 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_003680_230 |
0.0846830642 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 16 |
Hb_001077_030 |
0.084962831 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_143766_110 |
0.085203295 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 18 |
Hb_003633_070 |
0.0854108927 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 19 |
Hb_009112_010 |
0.0856028823 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 20 |
Hb_006615_120 |
0.0861066356 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |