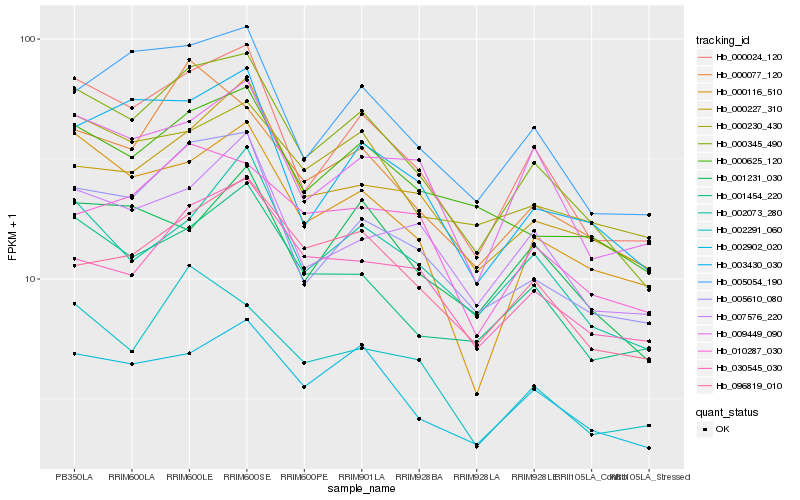
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_490 |
0.0 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 2 |
Hb_000024_120 |
0.0585954175 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 3 |
Hb_005610_080 |
0.0811120175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002902_020 |
0.0821647812 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 5 |
Hb_001454_220 |
0.091105062 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 6 |
Hb_000625_120 |
0.0911447097 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 7 |
Hb_005054_190 |
0.0912094241 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000077_120 |
0.0921508904 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 9 |
Hb_000116_510 |
0.096168834 |
- |
- |
PREDICTED: RNA-binding protein 24-A-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_003430_030 |
0.0972280899 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 11 |
Hb_002073_280 |
0.0984711935 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_096819_010 |
0.0994580005 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 13 |
Hb_002291_060 |
0.1002735494 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_000227_310 |
0.1021488224 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_009449_090 |
0.1028589033 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007576_220 |
0.1067465497 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 17 |
Hb_000230_430 |
0.1067829344 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_030545_030 |
0.1070612567 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 19 |
Hb_010287_030 |
0.1074897759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001231_030 |
0.1079322832 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |