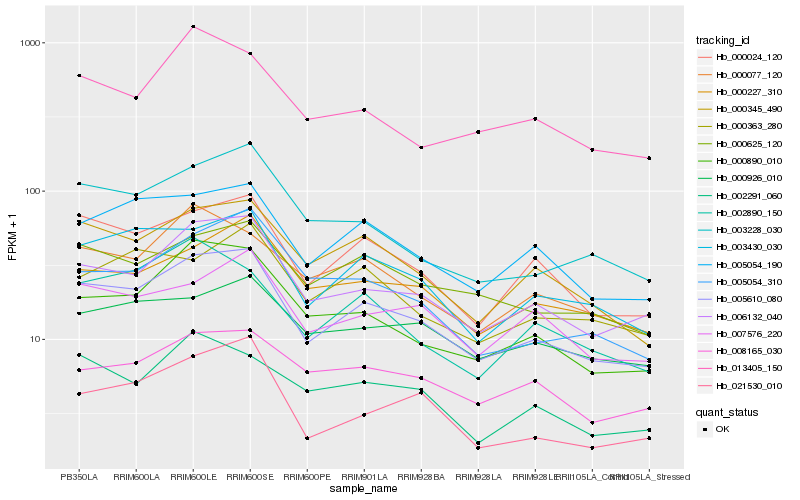
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005610_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006132_040 |
0.0697979895 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 3 |
Hb_000024_120 |
0.0736610958 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 4 |
Hb_003430_030 |
0.0775011544 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 5 |
Hb_000345_490 |
0.0811120175 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 6 |
Hb_005054_190 |
0.0845545495 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000890_010 |
0.0892451662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000625_120 |
0.0923797995 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 9 |
Hb_003228_030 |
0.0953683384 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 10 |
Hb_000227_310 |
0.0954471007 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000077_120 |
0.0983522156 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 12 |
Hb_007576_220 |
0.1049831809 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 13 |
Hb_021530_010 |
0.1052348488 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 14 |
Hb_005054_310 |
0.107813596 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 15 |
Hb_008165_030 |
0.1091099818 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 16 |
Hb_002291_060 |
0.1099034842 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_002890_150 |
0.1115438604 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000926_010 |
0.1120490536 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 19 |
Hb_013405_150 |
0.1125395737 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 20 |
Hb_000363_280 |
0.1129169651 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |