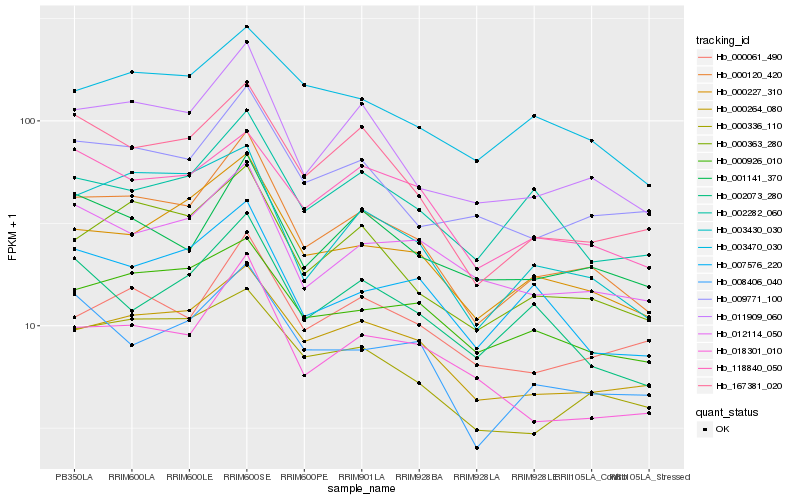
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_420 |
0.0 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 2 |
Hb_011909_060 |
0.0667604031 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 3 |
Hb_000363_280 |
0.0731568265 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 4 |
Hb_000227_310 |
0.0751629509 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000264_080 |
0.0792573914 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 6 |
Hb_167381_020 |
0.0864704275 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 7 |
Hb_003430_030 |
0.0895686106 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 8 |
Hb_000336_110 |
0.089950867 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_008406_040 |
0.0928883954 |
- |
- |
- |
| 10 |
Hb_018301_010 |
0.0932293836 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 11 |
Hb_009771_100 |
0.0948478745 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 12 |
Hb_118840_050 |
0.0954092556 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 13 |
Hb_012114_050 |
0.0964092693 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001141_370 |
0.0968634333 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 15 |
Hb_000061_490 |
0.1004137504 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 16 |
Hb_007576_220 |
0.1006288821 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 17 |
Hb_000926_010 |
0.100754436 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 18 |
Hb_002073_280 |
0.1026754819 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003470_030 |
0.1027104187 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 20 |
Hb_002282_060 |
0.1034887756 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |