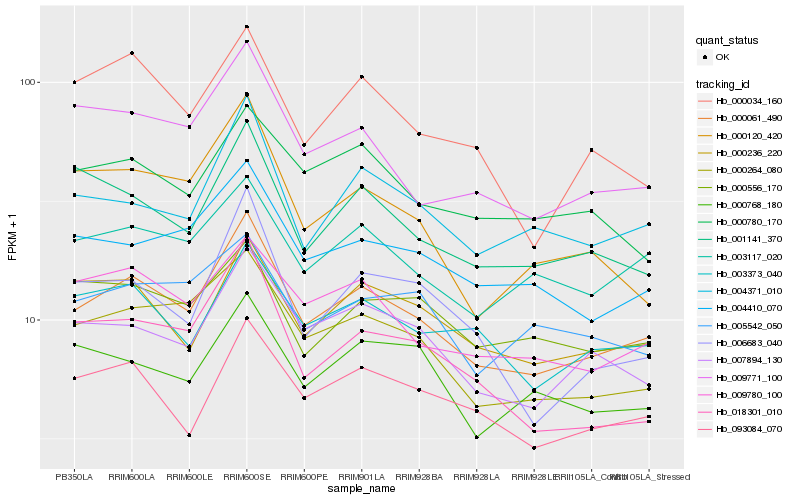
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_490 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 2 |
Hb_000264_080 |
0.0734796301 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 3 |
Hb_007894_130 |
0.0746541407 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 4 |
Hb_004371_010 |
0.0815285125 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_004410_070 |
0.0817268348 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 6 |
Hb_009780_100 |
0.0819155261 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 7 |
Hb_009771_100 |
0.0820696925 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 8 |
Hb_093084_070 |
0.0835597945 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 9 |
Hb_003117_020 |
0.0837881803 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 10 |
Hb_000236_220 |
0.0839104637 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 11 |
Hb_003373_040 |
0.0853395292 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 12 |
Hb_001141_370 |
0.0910203147 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 13 |
Hb_000780_170 |
0.0971401585 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 14 |
Hb_000768_180 |
0.0973719386 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 15 |
Hb_018301_010 |
0.0976933687 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 16 |
Hb_000034_160 |
0.0981075926 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 17 |
Hb_005542_050 |
0.0982624567 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 18 |
Hb_000556_170 |
0.0984817138 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 19 |
Hb_006683_040 |
0.0985993564 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 20 |
Hb_000120_420 |
0.1004137504 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |