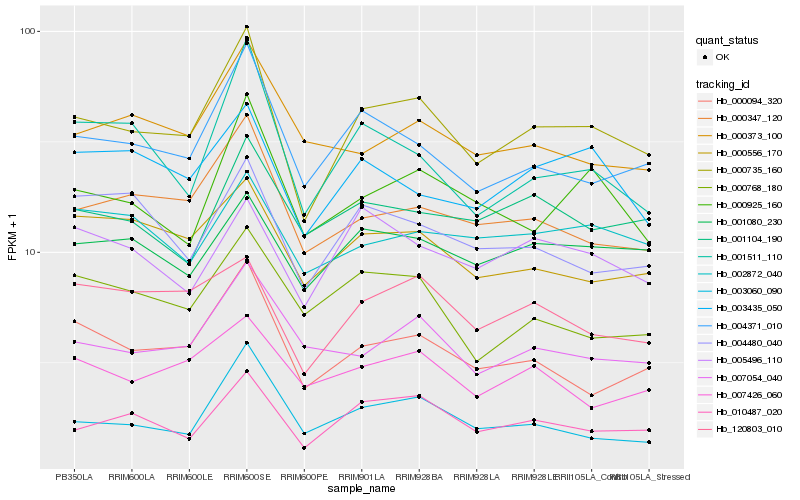
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000735_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 2 |
Hb_004371_010 |
0.0881829016 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 3 |
Hb_010487_020 |
0.0920215703 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 4 |
Hb_000347_120 |
0.0943835244 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 5 |
Hb_000094_320 |
0.098960718 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 6 |
Hb_000925_160 |
0.1037428598 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001080_230 |
0.1080176555 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 8 |
Hb_007054_040 |
0.1080666007 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 9 |
Hb_001511_110 |
0.1110519506 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_120803_010 |
0.1111880708 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein [Malus domestica] |
| 11 |
Hb_003435_050 |
0.1122214608 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1-like [Jatropha curcas] |
| 12 |
Hb_000556_170 |
0.1125846731 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001104_190 |
0.1171585186 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 14 |
Hb_003060_090 |
0.1181163924 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 15 |
Hb_007426_060 |
0.1183722506 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_000373_100 |
0.1187590566 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 17 |
Hb_000768_180 |
0.1193559286 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 18 |
Hb_004480_040 |
0.1197183463 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005496_110 |
0.1199175509 |
- |
- |
PREDICTED: uncharacterized protein LOC105107584 [Populus euphratica] |
| 20 |
Hb_002872_040 |
0.1203604772 |
- |
- |
pumilio, putative [Ricinus communis] |