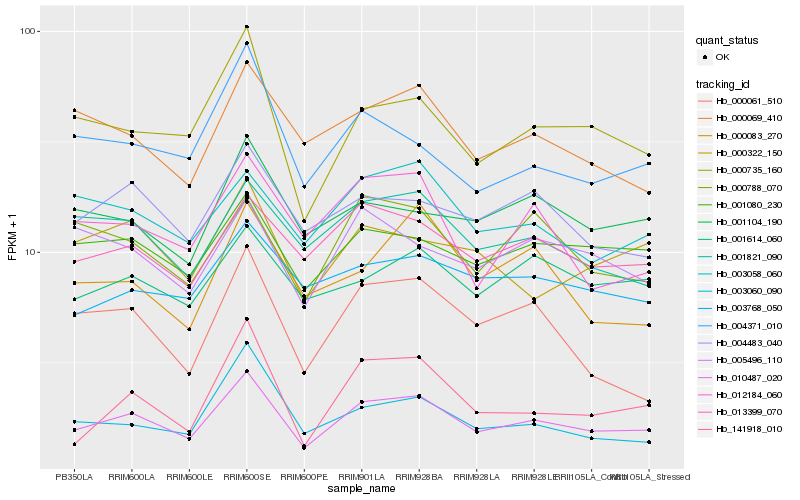
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010487_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 2 |
Hb_000735_160 |
0.0920215703 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 3 |
Hb_013399_070 |
0.1079660413 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001080_230 |
0.1131169546 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 5 |
Hb_004371_010 |
0.1134099411 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_001821_090 |
0.1142328569 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 7 |
Hb_004483_040 |
0.1146688092 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 8 |
Hb_000061_510 |
0.1152940972 |
- |
- |
hypothetical protein JCGZ_11686 [Jatropha curcas] |
| 9 |
Hb_000069_410 |
0.115329231 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 10 |
Hb_000788_070 |
0.1167406487 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 11 |
Hb_012184_060 |
0.1168407608 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003060_090 |
0.1175553333 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 13 |
Hb_141918_010 |
0.1190565858 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 14 |
Hb_003058_060 |
0.1193527325 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 15 |
Hb_001614_060 |
0.1204457512 |
- |
- |
hypothetical protein JCGZ_06918 [Jatropha curcas] |
| 16 |
Hb_000322_150 |
0.120552724 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003768_050 |
0.1220994435 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000083_270 |
0.1223189893 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001104_190 |
0.1223229566 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 20 |
Hb_005496_110 |
0.1223576368 |
- |
- |
PREDICTED: uncharacterized protein LOC105107584 [Populus euphratica] |