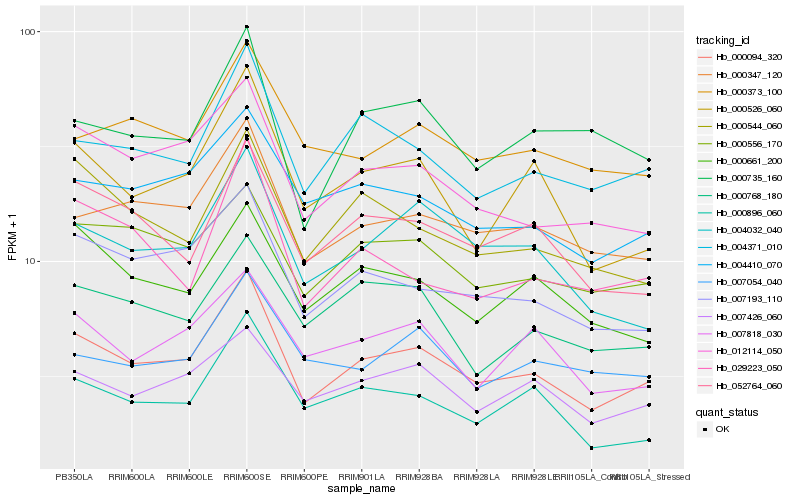
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_320 |
0.0 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 2 |
Hb_000347_120 |
0.0780734968 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 3 |
Hb_004371_010 |
0.0799147197 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 4 |
Hb_007193_110 |
0.0809097447 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 5 |
Hb_012114_050 |
0.0838370588 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000526_060 |
0.0865585319 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 7 |
Hb_004410_070 |
0.0879830932 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 8 |
Hb_004032_040 |
0.0893554724 |
- |
- |
unknown [Medicago truncatula] |
| 9 |
Hb_007426_060 |
0.0905546559 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_052764_060 |
0.0907845479 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 11 |
Hb_000556_170 |
0.0916225168 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000896_060 |
0.0918058254 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000544_060 |
0.0963112172 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 14 |
Hb_007818_030 |
0.0980922034 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 15 |
Hb_000735_160 |
0.098960718 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 16 |
Hb_000661_200 |
0.0996439575 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000373_100 |
0.1000126984 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 18 |
Hb_000768_180 |
0.1026401059 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 19 |
Hb_007054_040 |
0.1037510079 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 20 |
Hb_029223_050 |
0.1046768509 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |