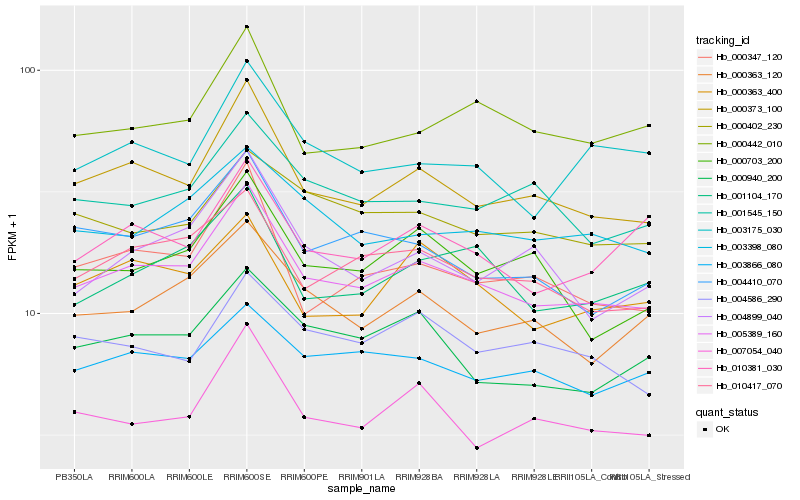
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005389_160 |
0.0 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 2 |
Hb_000373_100 |
0.0420591838 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 3 |
Hb_010417_070 |
0.0626765305 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000347_120 |
0.0677485342 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 5 |
Hb_000363_120 |
0.0704500147 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 6 |
Hb_001104_170 |
0.0713206607 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 7 |
Hb_003398_080 |
0.0737958659 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 8 |
Hb_007054_040 |
0.0745499844 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 9 |
Hb_001545_150 |
0.0751595972 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 10 |
Hb_003866_080 |
0.0752948634 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 11 |
Hb_000940_200 |
0.0763861926 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 12 |
Hb_000442_010 |
0.0764445572 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 13 |
Hb_004410_070 |
0.0767141535 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 14 |
Hb_000363_400 |
0.0788147665 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 15 |
Hb_000703_200 |
0.0795032533 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003175_030 |
0.079684442 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 17 |
Hb_000402_230 |
0.0810297434 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_010381_030 |
0.0820923483 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 19 |
Hb_004586_290 |
0.0825943286 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 20 |
Hb_004899_040 |
0.0826246444 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |