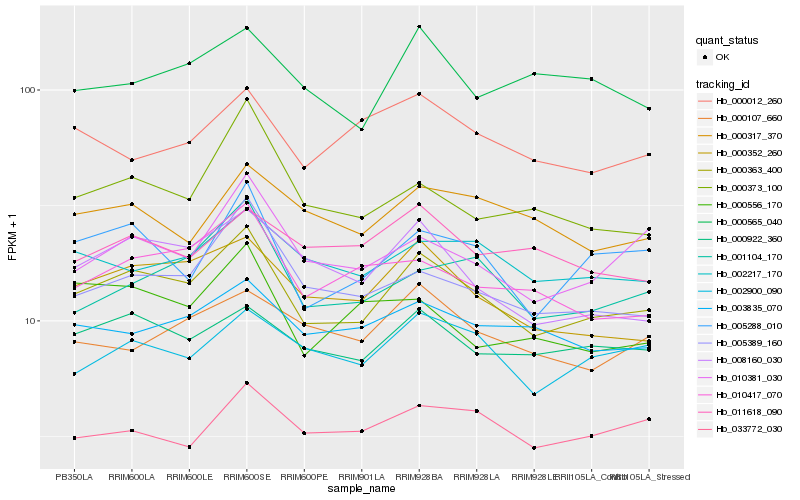
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_400 |
0.0 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 2 |
Hb_000352_260 |
0.0722701787 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 3 |
Hb_000922_360 |
0.07828676 |
- |
- |
- |
| 4 |
Hb_005288_010 |
0.0787754738 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_005389_160 |
0.0788147665 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 6 |
Hb_008160_030 |
0.0825150221 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_010381_030 |
0.0853395876 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 8 |
Hb_002900_090 |
0.0862651179 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 9 |
Hb_001104_170 |
0.0868016355 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 10 |
Hb_010417_070 |
0.0872680871 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002217_170 |
0.0878478588 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 12 |
Hb_003835_070 |
0.0899662416 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000373_100 |
0.0927623327 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 14 |
Hb_000317_370 |
0.0953538699 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 15 |
Hb_033772_030 |
0.0963063938 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 16 |
Hb_000565_040 |
0.0963368251 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 17 |
Hb_000107_660 |
0.0975840143 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 18 |
Hb_000012_260 |
0.0984410677 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 19 |
Hb_000556_170 |
0.0986187892 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 20 |
Hb_011618_090 |
0.09940828 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |