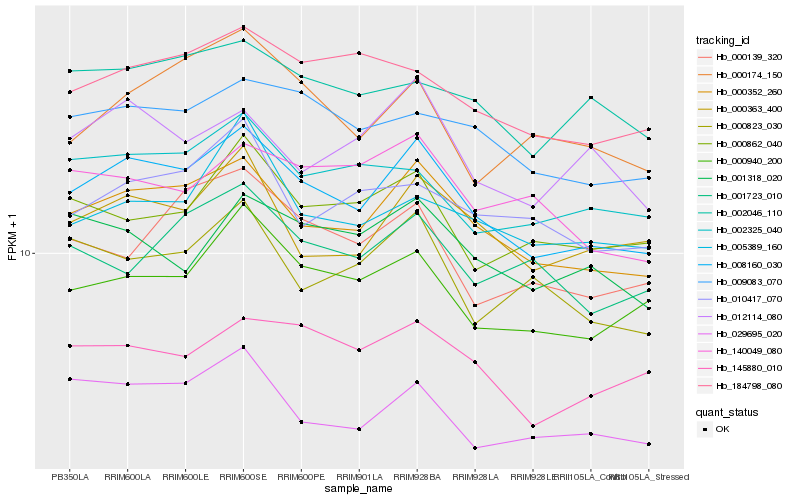
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008160_030 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000352_260 |
0.0387804142 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 3 |
Hb_000940_200 |
0.0802829982 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 4 |
Hb_009083_070 |
0.0816442459 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 5 |
Hb_000363_400 |
0.0825150221 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 6 |
Hb_145880_010 |
0.0875198587 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 7 |
Hb_184798_080 |
0.0899653967 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 8 |
Hb_002325_040 |
0.0909481188 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 9 |
Hb_029695_020 |
0.0911696913 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002046_110 |
0.0919522149 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001318_020 |
0.0940557174 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 12 |
Hb_000862_040 |
0.0948071809 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 13 |
Hb_012114_080 |
0.0954275943 |
- |
- |
gamma-glutamylcysteine synthetase [Hevea brasiliensis] |
| 14 |
Hb_000174_150 |
0.0957803122 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |
| 15 |
Hb_010417_070 |
0.096136277 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000139_320 |
0.0966701516 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 17 |
Hb_000823_030 |
0.0971697278 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 18 |
Hb_140049_080 |
0.0974174179 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 19 |
Hb_005389_160 |
0.0985824974 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 20 |
Hb_001723_010 |
0.099434097 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |