| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
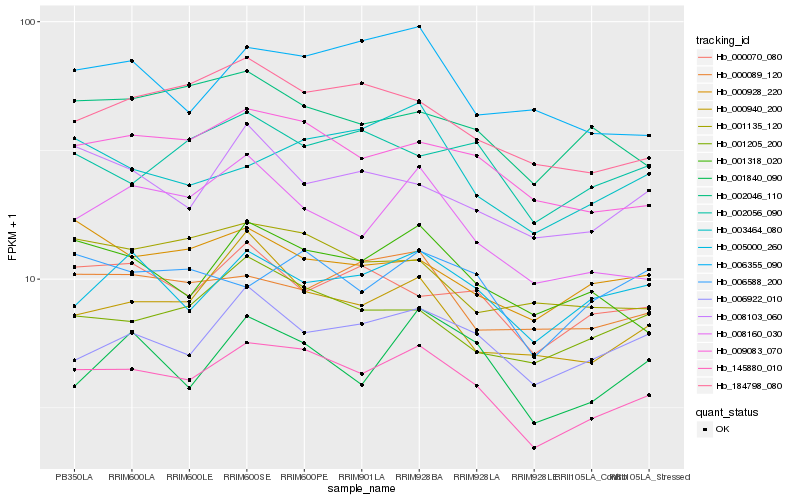
Hb_145880_010 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 2 |
Hb_009083_070 |
0.0715763025 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 3 |
Hb_001318_020 |
0.0835022441 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 4 |
Hb_184798_080 |
0.0872772198 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 5 |
Hb_006922_010 |
0.0875040811 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 6 |
Hb_008160_030 |
0.0875198587 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_001205_200 |
0.0890206881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000940_200 |
0.0907797254 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 9 |
Hb_005000_260 |
0.0915421568 |
- |
- |
recA family protein [Populus trichocarpa] |
| 10 |
Hb_006588_200 |
0.0921842065 |
- |
- |
3-beta-hydroxy-delta5-steroid dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000089_120 |
0.0929589441 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_002046_110 |
0.0937539111 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000070_080 |
0.0938252418 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003464_080 |
0.0940601188 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 15 |
Hb_001135_120 |
0.0961035056 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002056_090 |
0.096202965 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 17 |
Hb_000928_220 |
0.0968242443 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 18 |
Hb_006355_090 |
0.0972773456 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 19 |
Hb_008103_060 |
0.0975903155 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_001840_090 |
0.0978917633 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |