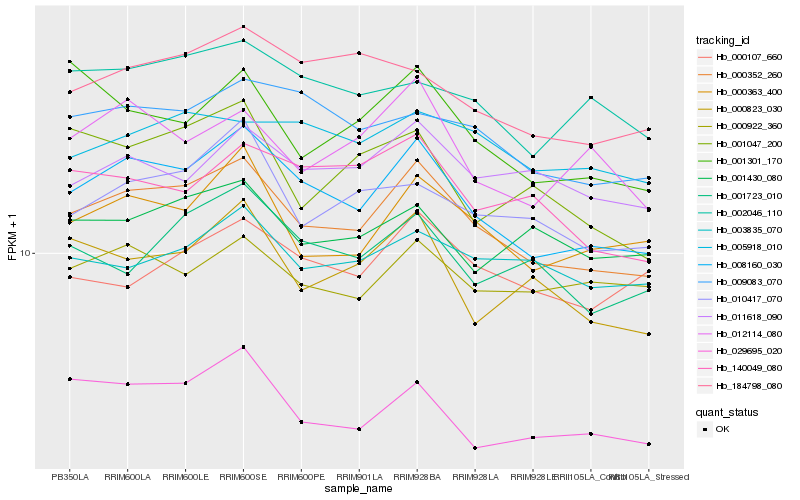
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_260 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 2 |
Hb_008160_030 |
0.0387804142 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000363_400 |
0.0722701787 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 4 |
Hb_010417_070 |
0.0832599368 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000823_030 |
0.0858099564 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 6 |
Hb_009083_070 |
0.0864000908 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 7 |
Hb_001723_010 |
0.0891505265 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 8 |
Hb_011618_090 |
0.0903647918 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_012114_080 |
0.0912079162 |
- |
- |
gamma-glutamylcysteine synthetase [Hevea brasiliensis] |
| 10 |
Hb_003835_070 |
0.0914443257 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000107_660 |
0.0915947305 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 12 |
Hb_140049_080 |
0.0918381054 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 13 |
Hb_002046_110 |
0.0923367528 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 14 |
Hb_184798_080 |
0.0930921161 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 15 |
Hb_001430_080 |
0.093093507 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 16 |
Hb_000922_360 |
0.0940834148 |
- |
- |
- |
| 17 |
Hb_001047_200 |
0.0943605389 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 18 |
Hb_001301_170 |
0.0944261058 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 19 |
Hb_005918_010 |
0.094434023 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 20 |
Hb_029695_020 |
0.0946780971 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |