| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
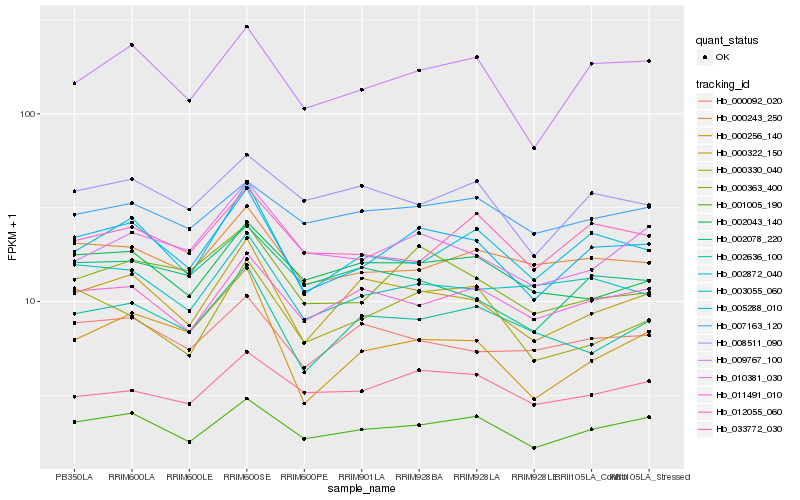
Hb_005288_010 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_009767_100 |
0.0625928094 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 3 |
Hb_001005_190 |
0.0657695144 |
- |
- |
PREDICTED: UPF0496 protein At3g19330-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000322_150 |
0.0767028632 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000363_400 |
0.0787754738 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 6 |
Hb_003055_060 |
0.0798256948 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002872_040 |
0.0909538053 |
- |
- |
pumilio, putative [Ricinus communis] |
| 8 |
Hb_011491_010 |
0.0916515269 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 9 |
Hb_000243_250 |
0.0916787824 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 10 |
Hb_010381_030 |
0.093242483 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 11 |
Hb_002636_100 |
0.094810213 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000256_140 |
0.0956592739 |
- |
- |
PREDICTED: uncharacterized protein LOC105637585 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002043_140 |
0.096749264 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 14 |
Hb_000330_040 |
0.0971233854 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 15 |
Hb_000092_020 |
0.0988932062 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_033772_030 |
0.1002395497 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 17 |
Hb_002078_220 |
0.1005386472 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008511_090 |
0.1010233034 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 19 |
Hb_012055_060 |
0.1011043893 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007163_120 |
0.1029016618 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |