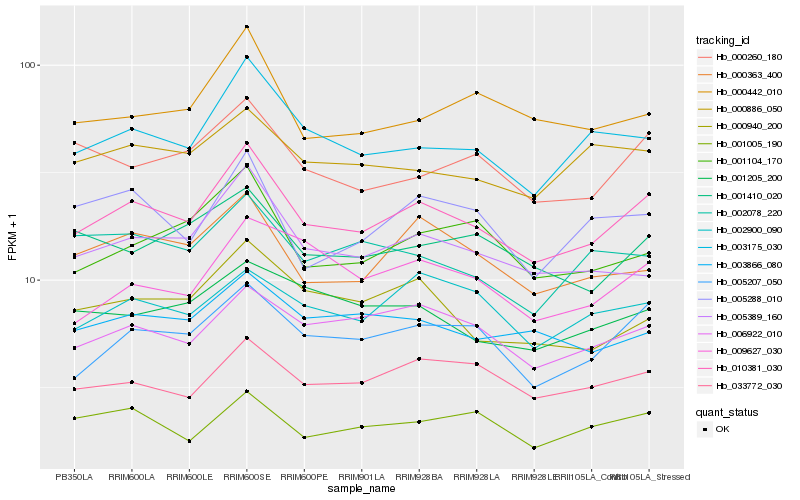
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010381_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 2 |
Hb_005207_050 |
0.0705495921 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 39 [Jatropha curcas] |
| 3 |
Hb_003175_030 |
0.0729547787 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 4 |
Hb_006922_010 |
0.0804401313 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 5 |
Hb_005389_160 |
0.0820923483 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 6 |
Hb_033772_030 |
0.0838514701 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 7 |
Hb_000363_400 |
0.0853395876 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 8 |
Hb_000260_180 |
0.0863614018 |
- |
- |
PREDICTED: KH domain-containing protein At2g38610 [Jatropha curcas] |
| 9 |
Hb_003866_080 |
0.0893246391 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 10 |
Hb_000940_200 |
0.0894201146 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 11 |
Hb_001104_170 |
0.0899255297 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 12 |
Hb_000886_050 |
0.0904729133 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 13 |
Hb_002900_090 |
0.091112237 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 14 |
Hb_000442_010 |
0.0924992975 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 15 |
Hb_001005_190 |
0.0930352347 |
- |
- |
PREDICTED: UPF0496 protein At3g19330-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_005288_010 |
0.093242483 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_001205_200 |
0.0938299059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_009627_030 |
0.093938061 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 19 |
Hb_001410_020 |
0.094030609 |
- |
- |
cdk8, putative [Ricinus communis] |
| 20 |
Hb_002078_220 |
0.0954715859 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |