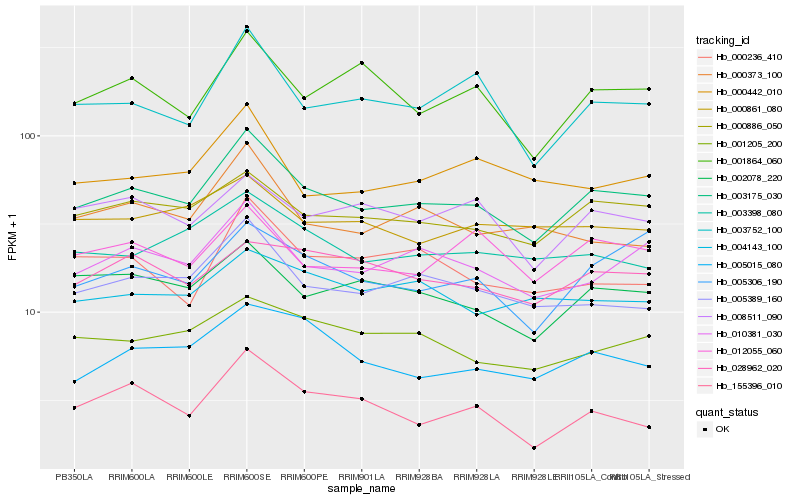
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003175_030 |
0.0 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 2 |
Hb_010381_030 |
0.0729547787 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 3 |
Hb_000886_050 |
0.0738396368 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 4 |
Hb_005389_160 |
0.079684442 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 5 |
Hb_002078_220 |
0.0833516806 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003752_100 |
0.084771981 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 7 |
Hb_003398_080 |
0.090107052 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 8 |
Hb_012055_060 |
0.0911244671 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001205_200 |
0.0925281856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005015_080 |
0.093028429 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001864_060 |
0.0932893457 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 12 |
Hb_005306_190 |
0.0948232854 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 13 |
Hb_008511_090 |
0.0957463724 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 14 |
Hb_000442_010 |
0.0957821417 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 15 |
Hb_000373_100 |
0.0977666584 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 16 |
Hb_000236_410 |
0.0987222107 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 17 |
Hb_000861_080 |
0.099171944 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 18 |
Hb_155396_010 |
0.099545072 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 19 |
Hb_028962_020 |
0.099846044 |
- |
- |
hypothetical protein POPTR_0016s00660g [Populus trichocarpa] |
| 20 |
Hb_004143_100 |
0.0999698559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |