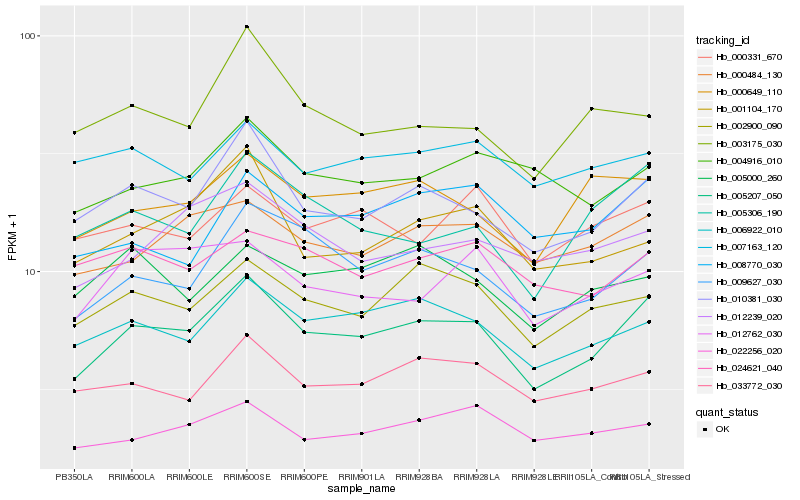
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005207_050 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 39 [Jatropha curcas] |
| 2 |
Hb_010381_030 |
0.0705495921 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 3 |
Hb_009627_030 |
0.0819630744 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 4 |
Hb_006922_010 |
0.0823656479 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 5 |
Hb_002900_090 |
0.0880493228 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 6 |
Hb_000484_130 |
0.0916402476 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 7 |
Hb_033772_030 |
0.0927316782 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 8 |
Hb_005306_190 |
0.0950372957 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 9 |
Hb_000649_110 |
0.0954073621 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 10 |
Hb_022256_020 |
0.1019380672 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35130, chloroplastic [Jatropha curcas] |
| 11 |
Hb_012239_020 |
0.1023049997 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 12 |
Hb_008770_030 |
0.1032888667 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290-like [Jatropha curcas] |
| 13 |
Hb_012762_030 |
0.1042180637 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 14 |
Hb_000331_670 |
0.1053082449 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 15 |
Hb_001104_170 |
0.1055158418 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 16 |
Hb_004916_010 |
0.1064282808 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 17 |
Hb_024621_040 |
0.1072871345 |
transcription factor |
TF Family: ERF |
hypothetical protein L484_014666 [Morus notabilis] |
| 18 |
Hb_005000_260 |
0.1086874607 |
- |
- |
recA family protein [Populus trichocarpa] |
| 19 |
Hb_003175_030 |
0.1090392524 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 20 |
Hb_007163_120 |
0.1091927302 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |