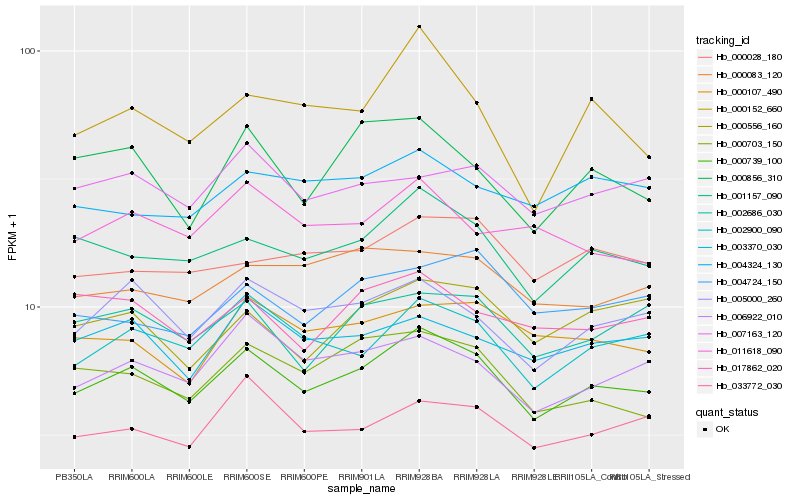
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000739_100 |
0.0 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 2 |
Hb_001157_090 |
0.0578605122 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 3 |
Hb_002900_090 |
0.0625777111 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 4 |
Hb_005000_260 |
0.0658842036 |
- |
- |
recA family protein [Populus trichocarpa] |
| 5 |
Hb_000703_150 |
0.0714565685 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A [Jatropha curcas] |
| 6 |
Hb_006922_010 |
0.072017169 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 7 |
Hb_002686_030 |
0.0752120617 |
- |
- |
PREDICTED: uncharacterized protein LOC105632635 isoform X1 [Jatropha curcas] |
| 8 |
Hb_033772_030 |
0.0752390161 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 9 |
Hb_003370_030 |
0.0768132513 |
- |
- |
PREDICTED: uncharacterized protein LOC105635547 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000107_490 |
0.0798841813 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 11 |
Hb_000856_310 |
0.0801269329 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 12 |
Hb_000083_120 |
0.0805641295 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_017862_020 |
0.0806498218 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 14 |
Hb_000028_180 |
0.0806642433 |
- |
- |
PREDICTED: probable serine/threonine protein kinase IREH1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007163_120 |
0.0811780048 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 16 |
Hb_000556_160 |
0.0813851223 |
- |
- |
PREDICTED: ankyrin repeat and zinc finger domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_000152_660 |
0.0825060318 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 18 |
Hb_011618_090 |
0.0831383461 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_004324_130 |
0.0849751821 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 20 |
Hb_004724_150 |
0.0850830182 |
- |
- |
expressed protein, putative [Ricinus communis] |