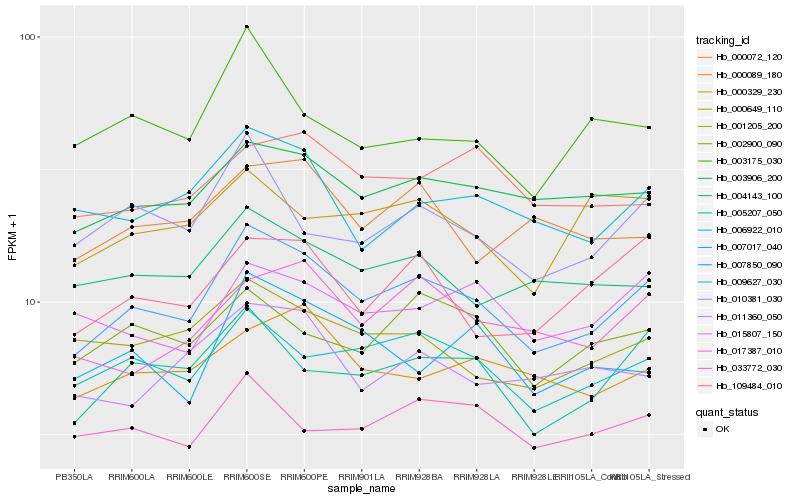
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009627_030 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 2 |
Hb_006922_010 |
0.0774651848 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 3 |
Hb_005207_050 |
0.0819630744 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 39 [Jatropha curcas] |
| 4 |
Hb_003906_200 |
0.0828350022 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 5 |
Hb_007017_040 |
0.0931432393 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 6 |
Hb_010381_030 |
0.093938061 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 7 |
Hb_001205_200 |
0.0942821377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004143_100 |
0.095141658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_017387_010 |
0.0953071986 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 10 |
Hb_002900_090 |
0.0968950761 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 11 |
Hb_033772_030 |
0.0971543105 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 12 |
Hb_109484_010 |
0.0972865657 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 13 |
Hb_000329_230 |
0.0981760857 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 14 |
Hb_015807_150 |
0.1004406854 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 15 |
Hb_011360_050 |
0.1025075838 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 16 |
Hb_007850_090 |
0.1027312401 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000089_180 |
0.1028610734 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 18 |
Hb_003175_030 |
0.1039455009 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 19 |
Hb_000072_120 |
0.1044833736 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000649_110 |
0.1045155947 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |