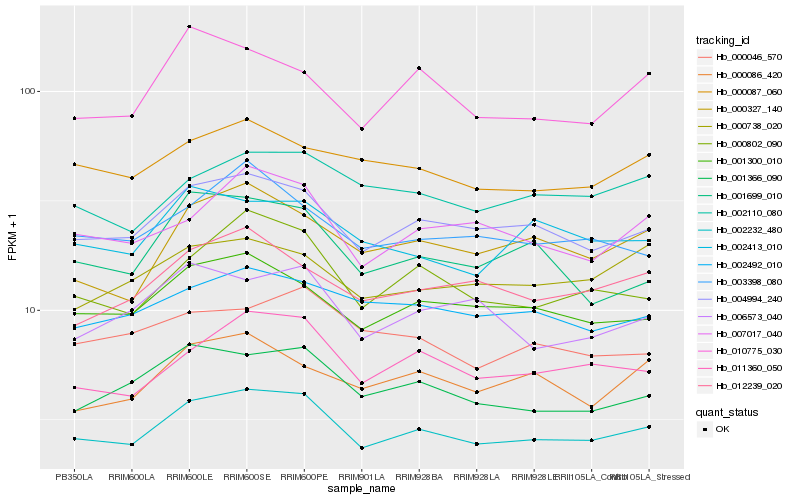
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_480 |
0.0 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 2 |
Hb_004994_240 |
0.0451677163 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 3 |
Hb_011360_050 |
0.0591010855 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000802_090 |
0.0608179044 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 5 |
Hb_001300_010 |
0.0631005285 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_001366_090 |
0.0668098758 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 7 |
Hb_007017_040 |
0.0689717082 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 8 |
Hb_000327_140 |
0.0733087365 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 9 |
Hb_012239_020 |
0.0748707482 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 10 |
Hb_003398_080 |
0.0759919568 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 11 |
Hb_002110_080 |
0.0764615612 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 12 |
Hb_002492_010 |
0.0767719385 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 13 |
Hb_000087_060 |
0.0773377494 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 14 |
Hb_002413_010 |
0.079334521 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_001699_010 |
0.0799631996 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 16 |
Hb_006573_040 |
0.0811035018 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_000086_420 |
0.08155797 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 18 |
Hb_010775_030 |
0.0821430511 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 19 |
Hb_000046_570 |
0.0827061699 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 20 |
Hb_000738_020 |
0.0827340867 |
- |
- |
zinc finger protein, putative [Ricinus communis] |