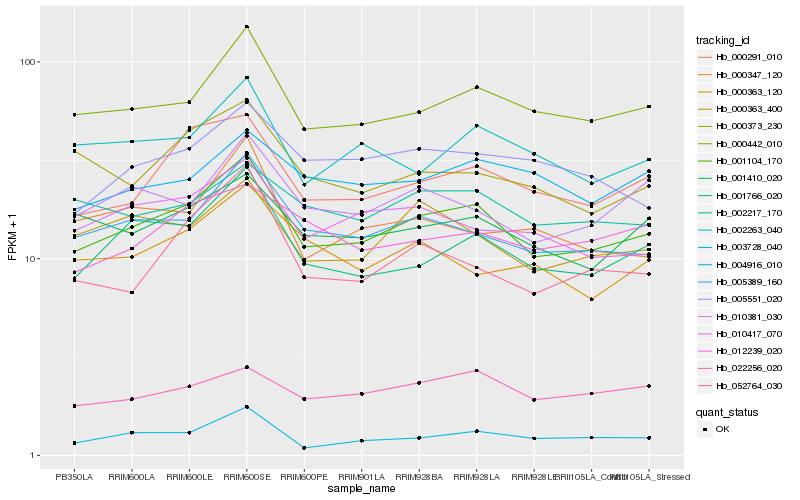
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001104_170 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 2 |
Hb_000442_010 |
0.0629663232 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 3 |
Hb_005389_160 |
0.0713206607 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 4 |
Hb_001766_020 |
0.0756700837 |
- |
- |
PREDICTED: probable galacturonosyltransferase 14 [Populus euphratica] |
| 5 |
Hb_000291_010 |
0.077561926 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 6 |
Hb_000363_400 |
0.0868016355 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 7 |
Hb_001410_020 |
0.0878521928 |
- |
- |
cdk8, putative [Ricinus communis] |
| 8 |
Hb_010417_070 |
0.0887655821 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002263_040 |
0.0888602121 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 10 |
Hb_010381_030 |
0.0899255297 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 11 |
Hb_022256_020 |
0.0939159292 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35130, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002217_170 |
0.0960337526 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 13 |
Hb_005551_020 |
0.0967531006 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 14 |
Hb_012239_020 |
0.0972358781 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 15 |
Hb_000373_230 |
0.0972376764 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 16 |
Hb_004916_010 |
0.0977202155 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 17 |
Hb_003728_040 |
0.0996597966 |
- |
- |
PREDICTED: uncharacterized protein LOC101492305 [Cicer arietinum] |
| 18 |
Hb_052764_030 |
0.0996614798 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 19 |
Hb_000347_120 |
0.0999153964 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 20 |
Hb_000363_120 |
0.1003954502 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |