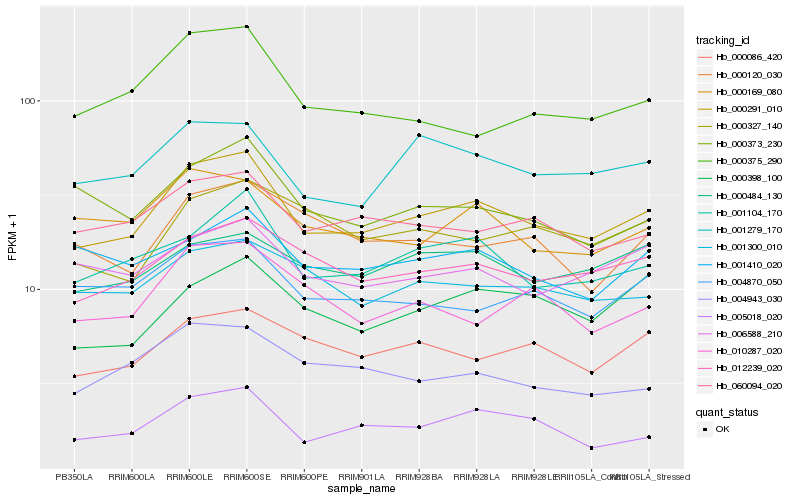
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_010 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 2 |
Hb_001104_170 |
0.077561926 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 3 |
Hb_012239_020 |
0.080399356 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 4 |
Hb_005018_020 |
0.08403777 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g07740, mitochondrial [Jatropha curcas] |
| 5 |
Hb_060094_020 |
0.0853012856 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 6 |
Hb_001279_170 |
0.0880412099 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000086_420 |
0.0918879801 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 8 |
Hb_000169_080 |
0.0927604509 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 9 |
Hb_001300_010 |
0.0935977508 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_004870_050 |
0.0955322417 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 11 |
Hb_006588_210 |
0.0960978843 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 12 |
Hb_010287_020 |
0.0965315328 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 13 |
Hb_000373_230 |
0.0968756663 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 14 |
Hb_004943_030 |
0.0975819671 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 15 |
Hb_000398_100 |
0.0990757401 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 16 |
Hb_000375_290 |
0.0998238773 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000484_130 |
0.1012443845 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 18 |
Hb_000120_030 |
0.102027999 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000327_140 |
0.1021503237 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001410_020 |
0.1025231136 |
- |
- |
cdk8, putative [Ricinus communis] |