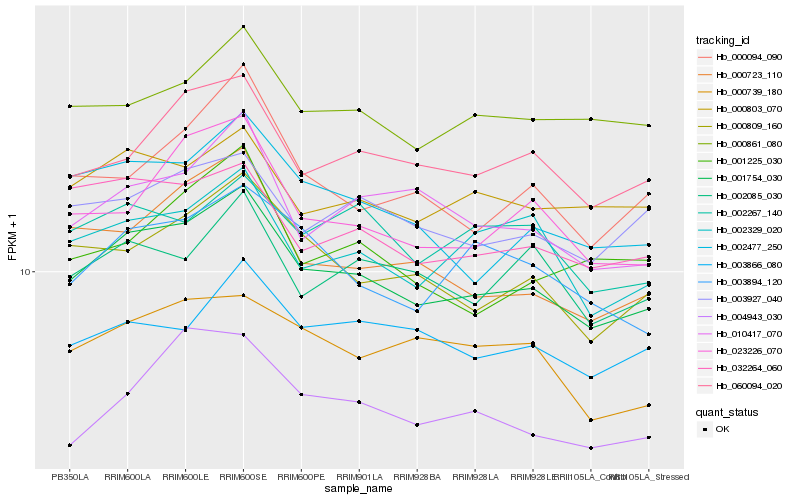
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_030 |
0.0 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 2 |
Hb_000809_160 |
0.0634385831 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 3 |
Hb_002477_250 |
0.0641635919 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 4 |
Hb_023226_070 |
0.0651802458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_060094_020 |
0.0653163208 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 6 |
Hb_032264_060 |
0.0673167296 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 7 |
Hb_000723_110 |
0.0682573434 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 8 |
Hb_000803_070 |
0.0712080896 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 9 |
Hb_010417_070 |
0.071262177 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000861_080 |
0.0720036341 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 11 |
Hb_002329_020 |
0.072599571 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 12 |
Hb_003927_040 |
0.0727130494 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 13 |
Hb_000094_090 |
0.0728407303 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003894_120 |
0.0731830775 |
- |
- |
PREDICTED: uncharacterized protein LOC105643110 [Jatropha curcas] |
| 15 |
Hb_003866_080 |
0.0746114464 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 16 |
Hb_004943_030 |
0.0772756631 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 17 |
Hb_002085_030 |
0.0772947381 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000739_180 |
0.0782135029 |
- |
- |
PREDICTED: uncharacterized protein LOC105643110 [Jatropha curcas] |
| 19 |
Hb_002267_140 |
0.0789390583 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 20 |
Hb_001225_030 |
0.0792437898 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |