| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
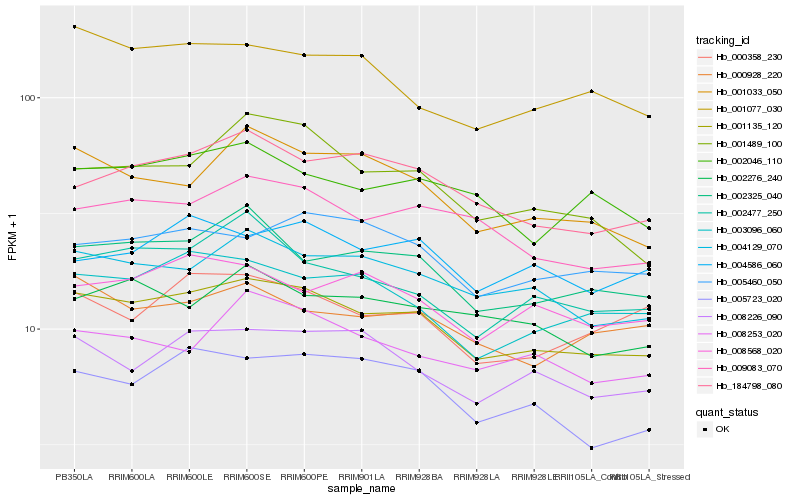
Hb_001135_120 |
0.0 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_004129_070 |
0.0567859599 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 3 |
Hb_002325_040 |
0.0568512103 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 4 |
Hb_009083_070 |
0.0629960435 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 5 |
Hb_003096_060 |
0.0642870484 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 6 |
Hb_001033_050 |
0.0662589439 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 7 |
Hb_184798_080 |
0.066968622 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 8 |
Hb_008226_090 |
0.0674242662 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002477_250 |
0.067539846 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 10 |
Hb_002046_110 |
0.0690483227 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005460_050 |
0.0698106573 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 12 |
Hb_005723_020 |
0.0704220656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000928_220 |
0.0709353845 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 14 |
Hb_008568_020 |
0.0716142365 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 15 |
Hb_001077_030 |
0.0718233749 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_008253_020 |
0.0725890794 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 17 |
Hb_001489_100 |
0.0739182187 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 18 |
Hb_002276_240 |
0.0744153908 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 19 |
Hb_000358_230 |
0.0759904916 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_004586_060 |
0.076247888 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |