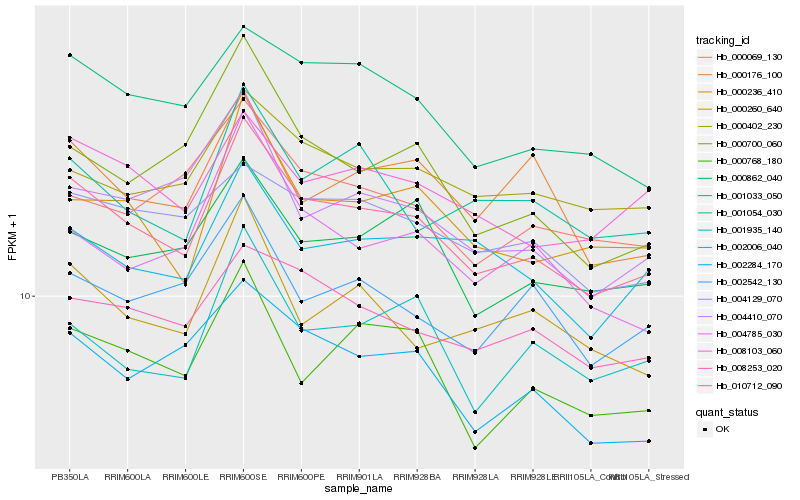
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010712_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 2 |
Hb_001033_050 |
0.0682692685 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 3 |
Hb_000236_410 |
0.0708738934 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 4 |
Hb_008253_020 |
0.0716292921 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 5 |
Hb_002284_170 |
0.0754975122 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 6 |
Hb_008103_060 |
0.0761172369 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 7 |
Hb_000862_040 |
0.0777490518 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 8 |
Hb_000402_230 |
0.0781590723 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002006_040 |
0.0782559662 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta isoform X1 [Jatropha curcas] |
| 10 |
Hb_000768_180 |
0.0795002815 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 11 |
Hb_004129_070 |
0.0803394975 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 12 |
Hb_001054_030 |
0.0804755045 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 13 |
Hb_004410_070 |
0.0818495343 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 14 |
Hb_002542_130 |
0.0818703299 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 15 |
Hb_004785_030 |
0.0822861703 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 16 |
Hb_000260_640 |
0.0825309003 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 17 |
Hb_000069_130 |
0.0825758762 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000700_060 |
0.083082075 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 19 |
Hb_001935_140 |
0.0835077819 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 20 |
Hb_000176_100 |
0.0843444593 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |