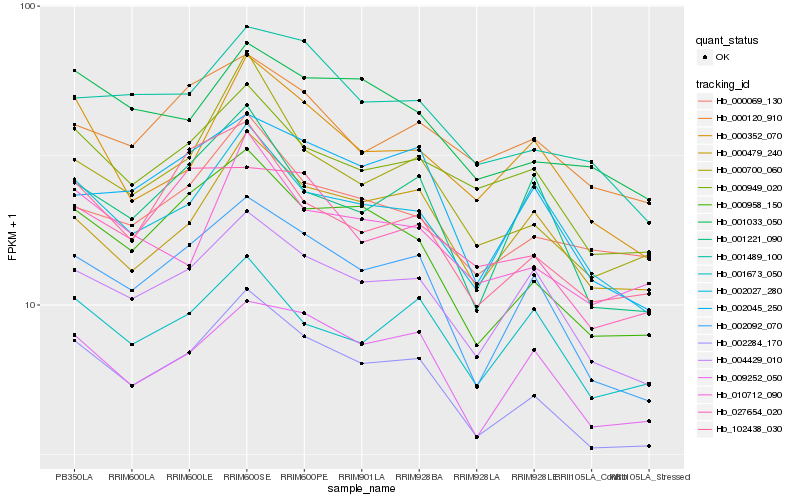
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_170 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 2 |
Hb_000958_150 |
0.0519525154 |
- |
- |
Actin, putative [Ricinus communis] |
| 3 |
Hb_002092_070 |
0.056820367 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 4 |
Hb_004429_010 |
0.0570035947 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_000949_020 |
0.0624420797 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 6 |
Hb_000700_060 |
0.0632823097 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 7 |
Hb_009252_050 |
0.0650766333 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_001033_050 |
0.0687736649 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 9 |
Hb_001489_100 |
0.0689521571 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 10 |
Hb_002027_280 |
0.0705311413 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 11 |
Hb_000069_130 |
0.071170164 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 12 |
Hb_102438_030 |
0.0712664194 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000479_240 |
0.0729830758 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002045_250 |
0.0745502142 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 15 |
Hb_010712_090 |
0.0754975122 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 16 |
Hb_000352_070 |
0.0766915942 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 17 |
Hb_001673_050 |
0.0783665535 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_027654_020 |
0.0784147666 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 19 |
Hb_000120_910 |
0.0784290141 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 20 |
Hb_001221_090 |
0.0789375662 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |